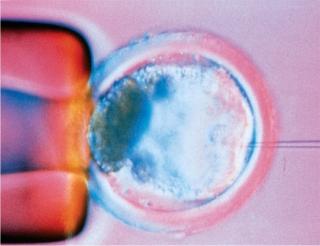
Every bioprocess begins with an expression system, and every expression system begins with DNA transfection. Derived from transformation and infection, the word paradoxically has come to be applied mainly to nonviral methods of genetically engineering cells; viral-vector–mediated DNA transfer is often called transduction. There are chemical, particulate, physical/mechanical, and viral means of getting new genetic material into a cell, and that DNA may take a number of different forms. Even the cloning method (pictured right) using a microscopic needle to inject a new cellular nucleus can be considered a form of transfection.
Short-lived pores can be opened up in the outer membranes of animal cells for uptake of genetic vectors such as supercoiled DNA plasmids or an artificial chromosomes. Calcium phosphate can be used to facilitate the movement of DNA across cellular membranes, as can cationic liposomes. Electroporation increases membrane permeability through the application of an electrical field, providing a physical rather than chemical means to introduce new genes to cells. Viruses, of course, have evolved specifically to force their way in and introduce their own foreign DNA to hijack cellular machinery for reproducing themselves in large quantities. So they can also be engineered for infecting and transforming cells. Some less commonly used techniques involve sonic poration, tiny lasers, dendrimers, magnetic forces, cationic polymers, and gene guns.
A number of suppliers offer a broad range of transfection reagents for sale: e.g., Agilent Technologies, Altogen Biosystems, Clontech, EMD Millipore, GeneCopoeia, Invitrogen (Life Technologies), Mirus Bio, New England Biolabs, Origene, Polyplus Transfection, Promega, Qiagen, Roche Applied Science, Sigma-Aldrich, SignaGen Laboratories, and Thermo Scientific. The array of options seems to be endless, but as with many other aspects of bioprocessing, that is a reflection of the biological complexities inherent in this industry. What works with one product/process might not work with another — in fact, it probably won’t.
Rather than introducing new genes, another transfection method is used to suppress them with small interfering ribonucleic acid (siRNA) sequences. This technology, however, has found more utility in the research world than in biopharmaceutical development — although some companies are exploring siRNA molecules as potential therapeutics, themselves.
Brief History
The history of DNA transfection is the history of biotechnology itself. In 1967, a few research groups working independently discovered DNA ligases (1). In 1970, Hamilton Smith’s laboratory at the University of California (Berkeley) discovered restriction enzymes, enabling scientists to isolate genes from an organism’s genome using gel electrophoresis. Once both were understood, it would be possible to “cut and paste” DNA sequences. And in 1972, Paul Berg did just that at Stanford University in northern California to create the first recombinant DNA molecules (virus-based), for which he later won a 1980 Nobel Prize in chemistry.
Around the same time, Herbert Boyer (later a cofounder of Genentech) and Stanley Cohen met at a conference on bacterial plasmids (2). Boyer’s laboratory at the University of California (San Francisco) had isolated an enzyme that could be used to cut DNA precisely — while not too far away at Stanford University, Cohen had developed methods for isolating and cloning genes and introducing new plasmids into bacteria. (Plasmids had been identified by Joshua Lederberg at the University of Wisconsin—Madison in 1952.) Four months of collaboration using those complementary techniques led to what we know of as genetic engineering and the creation of the world’s first genetically modified organism: bacteria that survive in the presence of kanamycin (2). Since then, the technology has been refined by discovery of new enzymes to cut DNA differently and by mapping of the genetic code of different organisms.
Expanding Methods
A plasmid can carry about 30–40 kbp of introduced DNA. Later developments gave the burgeoning biopharmaceutical industry more options for DNA transfection: e.g., artificial chromosomes, supercoiled DNA, and zinc-finger nucleases (ZFNs). Bacterial artificial chromosomes (BACs) are based on fertility plasmids (F-plasmids) that contain partition genes to promote even distribution of plasmids following bacterial cell division. A BAC can introduce 150–350 kbp. Yeast artificial chromosomes (YACs) are vectors for 100- to 3,000-kbp DNA fragments, and they offer a eukaryotic advantage over BACs. YACs can be used to express eukaryotic proteins that require posttranslational modification, but their large size makes them less stable. There are also yeast-plasmid vectors. Mammalian artificial chromosomes (MACs) are even larger, able to transfer several thousand base-pairs of DNA, but they can be made more stable using complex engineering techniques such as the ACE system (3).
By over- or underwinding the strands within cells, supercoiling naturally compacts DNA, which can be thousands of times longer than a cell itself, especially during cell division. Enzymes such as topoisomerase can be used to supercoil DNA and facilitate transcription. DNA must be unwound for polymerase action, so the region ahead of a polymerase complex will be unwound, and the resulting physical stress is ameliorated by positive supercoils beyond. Behind the complex, DNA is rewound, so negative supercoils must compensate. Plasmid structure and shape have proven to be important parameters for determining transfection efficiency. Covalently closed, circular (supercoiled) DNA adopts a compact form that is optimal for transfection efficiency. Companies such as Althea Technologies (San Diego, CA) and the PlasmidFactory (Bielefeld, Germany) are exploiting control of this phenomenon.
Zinc-finger nucleases (ZFNs) are artificial restriction enzymes created in the 1990s by scientists who fused a zinc-finger DNA-binding domain with a DNA-cleavage domain. The former can be engineered to target desired DNA sequences, enabling ZFNs to target unique sequences within complex genomes. These transcription reagents take advantage of natural DNA repair machinery to precisely alter the genomes of higher organisms.
Transfection in the BPI Archives at WWW.BIOPROCESSINTL.COM
Kunert R, et al. Control of Key Parameters in Developing Mammalian Production Clones. BioProcess Int. 2(6) 2004: 54–59.
Julien C. Production of Humanlike Recombinant Proteins in Pichia pastoris. BioProcess Int. 4(1) 2006: 22–32.
Ludwig DL. Mammalian Expression Cassette Engineering for High-Level Protein Production. BioProcess Int. 4(5) 2006: S14–S23.
Kim HY. Improved Expression Vector Activity Using Insulators and Scaffold/ Matrix-Attachment Regions. BioProcess Int. 4(5) 2006: S24–S31.
Kayser K, et al. Cell Line Engineering Methods for Improving Productivity. BioProcess Int. 4(5) 2006: S6–S14.
Kline JB,
et al. Whole-Genome Evolution Technology. BioProcess Int. 4(5) 2006: S42–S47.
Shi J, Yang J. Transient Gene Silencing in NS/0 Suspension Cell Culture By siRNA. BioProcess Int. 5(9) 2007: 72–77.
Kunert R, Gach J, Katinger H. Expression of a Fab Fragment in CHO and Pichia pastoris. BioProcess Int. 6(6) 2008: S34–S40.
Liu X, et al. Isolation of Novel High- Osmolarity Resistant CHO DG44 Cells After Suspension of DNA Mismatch Repair. BioProcess Int. 8(4) 2010: 68–76.
Girod P-A, et al. Rapid Production of Functional Proteins of a Combinatorial IgG Library in CHO Cells. BioProcess Int. 10(1) 2012: 58–61.
Ausubel LJ, et al. Production of CGMP-Grade Lentiviral Vectors. BioProcess Int. 10(2) 2012: 32–43.
Agrawal V, et al. A High-Yielding, CHO-K1–Based Transient Transfection System. BioProcess Int. 11(1) 2013: 28–35.
In 2011, SAFC (St. Louis, MO) licensed the technology from Sangamo Biosciences (Richmond, CA) for use in engineering Chinese hamster ovary (CHO) cells for bioproduction. This is a perfect example of the legal complexities found in the modern practice of gene transcription. In the four decades that have passed since the first DNA recombinations, the gene sequences associated with different expression vectors and systems have become some of the most valuable intellectual property that biotechnology companies possess.
Applications
When you consider the biopharmaceutical applications of gene transfection, cell-line development is of course the first that comes to mind. Protocols vary by reagent and cell line, but some basic rules apply across the board. The first question to answer is whether you’ll be transfecting for stable or transient expression; the latter is increasingly used as a means of producing testable material faster in early product development stages. For transient expression, sometimes a genetically engineered virus induces expression (as in the baculovirus system with insect cells), and sometimes cells are transfected directly. They are typically harvested 24–72 hours afterward (4).
The goal of stable, long-term transfection is to isolate and propagate individual clones containing transfected DNA that has integrated into the cellular genome. Distinguishing nontransfected cells from those that have taken up exogenous DNA involves selective screening. This screening can be accomplished by drug selection when an appropriate drug-resistance marker is included in the transfected DNA. Alternatively, morphological transformation can be used as a selectable trait in certain cases. For example, bovine papilloma virus vectors produce a morphological change in transfected mouse CI127 cells. (4)
Cells are assayed to determine the success of transfection, depending on the marker or reporter gene chosen. Reporter gene assays are usually performed 1–3 days after transfection, often using luminescence detection. Green fluorescent protein (GFP) is a common reporter. Some people may choose a dual-reporter system to help them better determine the efficiency of their transfection process.
Factors that influence transfection efficiency range from the health and size/density of the cell population used, the quantity and quality of DNA and transfection reagents used (no proteins, RNA, or other contaminants), and the number of culture passages (which should be kept below 50). “With any transfection reagent or method used, some cell death will occur” (4). As in any bioprocess operation, optimization work to find the best possible design space for the transfection process will be rewarded with notable success: e.g., specifying the best charge ratio for cationic reagents, the amount of DNA required, the length of time for cells to be exposed to reagents, and the kind of culture medium that will be used.
Transfection methods and options continue to evolve as new products and methods are launched every year “with improved efficiency and less cytotoxicity” (5). As our May 2006 supplement on cell-line engineering pointed out (www.bioprocessintl.com/journal/supplements/2006/May), many special promoters and other gene sequences have helped improve transfection efficiencies and resulting expression systems for higher and higher product titers in biopharmaceutical production.
Until recently, most cell line development procedures were based on random integration and gene amplification, but several methods for targeted genetic modification of cells have been developed. Some of those are homologous recombination, RNA interference and zinc-finger nucleases. Especially the latter two have evolved considerably and will soon become a standard for cell line engineering in research and industrial application. (6)
Experts see the technology moving toward increased precision, drilling down to deliver RNA to specific cellular organelles — and even potentially expanding beyond cell lines to whole organisms (5, 6).
And that brings me to the other biotherapeutic applications of DNA transfection: gene and cell therapies. “Safe and reliable transfection methods that can be applicable to humans are needed to establish clinical therapeutics” (5). Many methods of transfection that work just fine in cell culture are out of the question for delivering genes to humans as therapeutics (7). Plasmid, oligonucleotide, DNA-based ribozymes, aptamers, and even siRNAs commonly involve viral, polymer, or liposomal delivery systems. But they are plagued by low transfection efficiency as well as immunogenicity and other difficulties. Although many gene therapies have been in development, very few have made it to market anywhere in the world. For the subset of cell therapies that involve genetic manipulation, the transfection process occurs outside patients and thus follows the general approach of other cell-culture–based methods.
Getting Together
DNA transfection is such an integral part of biotechnology that it doesn’t really get its own conference focus, although you’ll see it as a subtheme in many gene-therapy conferences such as
Phacilitate’s annual “Cell and Gene Therapy Forum” (which was held this past January in Washington, DC, www.cgt-forum.com)
the annual meeting of the American Society of Gene and Cell Therapy (15–18 May 2013 in Salt Lake City, www.asgct.org)
the annual conference of the British Society of Gene and Cell Therapy (March 2014 in London, www.bsgct.org)
Cell-line engineering is also a subject for which gene transfection comes heavily into play. Preeminant among these is IBC Life Sciences’ annual “Cell Line Development and Engineering,” which is 20–22 May this year in La Jolla, CA (www.ibclifesciences.com/CellLine). The company produced its third such event for Asia in Shanghai, China, this past March — and we expect to see more installments of that one in the future (www.celllineasia.com). Engineering Conf
erences International presents “Cell Culture Engineering XIV” (a biennial event) next May in Quebec City, Canada. Terrapin’s “Cell Culture World Congress USA” will be this fall in Cambridge, MA (www.terrapinn.com/2013/cell-culture-world-congress-usa), and its “World Cell Culture Congress” will be in Munich, Germany, for February of 2014. Finally, Oxford Global Conferences will present its second annual “Cell Culture and Bioprocessing Congress” this November in London (www.cellculture-congress.com).
About the Author
Author Details
Cheryl Scott is cofounder and senior technical editor of BioProcess International, 1574 Coburg Road #242, Eugene, OR 97401; 1-646-957-8879; [email protected].
REFERENCES
1.) Kiermer, V. 2007.The Dawn of Recombinant DNA Nature Meth.
2.) 1997.Inventor of the Week: Cloning of Genetically Engineered Molecules Lemelson-MIT, Massachusetts Institute of Technology, Boston.
3.) Lindenbaum, M. 2004. A Mammalian Artificial Chromosome Engineering System (ACE System) Applicable to Biopharmaceutical Protein Production, Transgenesis and Gene-Based Cell Therapy. Nucl. Acids Res. 32:e172.
4.) 2011.Chapter 12: Transfection Protocols and Applications Guide, Promega Corporation, Madison.
5.) Kim, TK, and JH. Eberine. 2010. Mammalian Cell Transfection: The Present and the Future. Analyt. Bioanalyt. Chem. 397:3173-3178.
6.) Krämer, O, S Klausing, and T. Noll. 2010. Methods in Mammalian Cell Line Engineering: From Random Mutagenesis to Sequence-Specific Approaches. Appl. Microbiol. Biotechnol. 88:425-436.
7.) Patil, SD, DG Rhodes, and DJ Burgess. 2005. DNA-Based Therapeutics and DNA Delivery Systems: A Comprehensive Review. AAPS J. 7:E61-E77.