From rapidly obtaining sufficient amounts of active protein in early stage development to cost effectively producing kilogram and even metric ton quantities for commercial supply, protein expression is critical at every stage of biopharmaceutical drug development. Having a high-performance protein expression platform across all stages is invaluable for the speed and success of protein and vaccine development.
Historically, biopharmaceutical researchers and process development scientists have used Escherichia coli in their laboratories to generate small quantities of protein. If target expression was low or insoluble, they undertook larger-volume fermentation or performed a refolding step. If a product could not be expressed by E. coli, they would investigate alternative hosts (e.g., yeast, insect or mammalian cells). This iterative and linear process is both time consuming and expensive.
Dowpharma’s Pfēnex expression technology platform addresses challenges of speed and cost at every phase of development. It enables parallel analysis of hundreds of expression strains at the beginning of preclinical development — combined with a scalable, high-density fermentation process that uses an inexpensive, completely defined medium.
The History
The Pfēnex platform is based on Pseudomonas fluorescens, strain MB101, biovar I which was isolated from a lettuce leaf in San Diego in the early 1980s by scientists at Mycogen Corporation. Initially, it was used for large-scale production of insecticidal proteins for the control of plant disease. That early application demonstrated its ability to achieve high cell densities using minimal medium and to produce large amounts of protein at very low cost. After aquiring Mycogen in 1998, The Dow Chemical Company invested significantly in R&D to further enhance the overall flexibility and performance of this strain for large-scale production of many industrial enzymes.
In early 2002, a team of scientists from Dow’s corporate R&D group joined with personnel from its biopharmaceutical contract manufacturing business to explore potential applications of P. fluorescens for production of therapeutic proteins and vaccine antigens. During the subsequent two years, that multidisciplinary team followed a systematic process to develop and commercialize the technology platform. Ultimately it was launched in January 2004 at the Informex trade show in Las Vegas, NV.
Platform and Toolbox Foundation
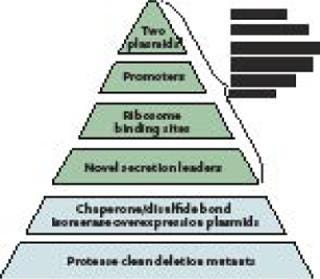
The Pfēnex expression technology platform was designed to assess performance of many host-strain–plasmid combinations in parallel to rapidly identify the optimal choice for high-yield expression of soluble, active target protein. The platform is underpinned by the sequence of the entire P. fluorescens genome. That knowledge allowed Dow to implement directed genetic modifications rapidly, facilitating sophisticated pathway engineering and enhancement of specific gene expression. Combining the annotated genome and functional genomics technologies (DNA microarrays and proteomics) the Dow team constructed a large library of P. fluorescens host strains with phenotypes designed to enhance the quality and stability of recombinantly expressed proteins (Figure 1).
Figure 1: ()
Genetic Elements
Plasmids: Having two compatible plasmids allows for induction of “helper proteins” (such as disulfide bond isomerases) before induction of a target protein gene encoded on a second plasmid. This ensures that conditions within the periplasm are ideal for folding once a target protein is expressed and secreted into it. These plasmids are stably maintained through complementation, rather than by selection for drug resistance, which removes the need for antibiotics in the process (1).
Promoters: Typically the tac promoter adapted from E. coli is used to drive expression of target genes. Native P. fluorescens promoters (2) have been engineered to drive accessory functions (e.g., chaperone expression). These promoters are inducible by small molecules (e.g., mannitol, benzoate) to prevent downstream processing issues.
Ribosome Binding Sites: Factors such as transcription and translation rates have been shown to affect the amount and quality of protein expressed. Having several ribosome binding site options can enhance the quality of the protein being expressed (3). The Pfēnex platform uses a range of ribosome binding sites on preconstructed plasmids to modulate translation initiation.
Secretion Leaders: Because disulfide bonds are structurally important to many therapeutic proteins (e.g., monoclonal antibodies and fragments), several P. fluorescens-native, sec-type secretion leaders have been developed (4). These direct localization of expressed protein to the subcellular compartment (periplasm), where formation of such bonds is catalyzed. Removal of the secretion leader occurs in vivo through a signal peptidase present on the inner cell membrane. Secretion into the P. fluorescens periplasm has proven to be a robust and efficient process that does not reduce expression level. Simply changing the secretion leader while keeping all other elements constant also significantly affects protein expression, so a large number of secretion leaders have been fully characterized and added to the Pfēnex toolbox.
Host Strains
Chaperone/Disulfide-Bond Isomerases: The most difficult issue in expression of mammalian, disulfide-bonded proteins in bacteria is misfolding and accumulation of insoluble aggregates. Bacteria can fold target proteins using oxidoreductases and isomerases present in the periplasm. Many reports show that overexpression of such enzymes can aid in expression of properly folded target proteins. Unfortunately, for every reported success, there seems to be a case that didn’t work. So the Dow team assembled a large set of host strains that can regulate overexpression of native P. fluorescens enzymes that influence recombinant protein folding. Dozens of such strains are in the toolbox of hosts available for high-throughput screening.
Protease Deletion Strains: Protein stability is another roadblock to efficient product expression. Degradation or partial degradation of expressed proteins not only can hurt overall yield (5), but also creates serious downstream purification issues. Dow has engineered dozens of protease deletion strains, many regularly induced upon recombinant protein expression. Single, complete gene deletion strains (and many multiple deletion strains) are available for high-throughput screening.
Recently Dow assembled a set of more than 100 unique expression vectors that combine different transcription, translation, and secretion signals. These can be used to express either one or two polypeptides.
Predicting which parts of the toolbox to use is crucial in successful expression of any given protein. This cannot be determined from intrinsic information (e.g., amino-acid sequence). Combining critical factors is empirical for each
target protein. Accordingly, Dow developed a high-throughput screening process that enables parallel evaluation of hundreds of unique host strains containing a number of expression strategies for a specific gene. When designing a strain engineering and screening strategy, scientists evaluate certain attributes of a protein to be expressed (e.g., molecular weight, propensity for proteolytic degradation, presence of cysteine residues, and expression performance in other hosts). For example, if a protein contains several disulfide bonds and is prone to proteolysis, a plasmid engineering strategy would typically include evaluation of several secretion leaders. Those plasmids would be transformed into a set of protease knockout and folding modulator overexpression strains.
Strains are initially screened at 96-well, 0.5-mL scale that exploits the obligately aerobic nature of P. fluorescens. Organic acids, acetate, and pyruvate do not accumulate when an obligate aerobic culture becomes oxygen limited. Such process byproducts are typically detrimental to culture health and are obstacles to efficient scale-up, a key difference between P. fluorescens and E. coli. Pfenex expression technology can achieve cell densities of 30–50 OD units at the 0.5-mL scale, which can generate sufficient protein for early analyses (e.g., N-terminal sequencing, bioactivity, and even small-scale purification). Similar experiments using E. coli in a defined medium typically achieve ∼5 OD units of cells and consequently much less biomass and target protein to evaluate.
Following strain screening, Dow undertakes a fermentation range-finding process. Selected strains are evaluated in 1-L fermentors under a range of growth and gene induction conditions. The results can be used to design the elements of an effective fermentation process. Additionally, these experiments yield sufficient biomass and target protein to begin primary recovery, purification, and protein characterization steps. At this stage, a three- to fivefold or higher improvement in expression level is typically observed. That reflects an increase in both biomass and specific protein expression frequently found under controlled, high–cell-density fermentation conditions used at this scale.
Regulatory Expectations
Regulatory expectations were taken into account when the Pfēnex platform was adapted for production of therapeutic proteins and vaccines. Pathogenicity and toxicology studies concluded that the organism is safe for such uses. The American Type Culture Collection (ATCC) has designated strains of P. fluorescens MB101 biovar I under biosafety level 1 (BSL-1), which defines them as “having no known potential to cause disease in humans or animals.”
Rather than antibiotic-resistance selection, Dow uses auxotrophic complementation to maintain plasmids, which has proven to be a very robust approach (1). A fully defined mineral-salts medium is used during strain development, process development, and scale-up — and it is completely free of animal components and organic nitrogen. Purification methods currently used for other gram-negative hosts can be used for proteins expressed by P. fluorescens. Dow collaborated with Cygnus Technologies to develop a fully validated host-cell–protein kit that became commercially available through Cygnus in April 2007.
Case Studies
Multigram-per-Liter Expression of a Fab Fragment: Antibodies and antibody derivatives are particularly difficult to express in prokaryotic systems because of their complex nature (containing multiple polypeptide chains). They contain multiple disulfide bonds, so they must be secreted into periplasm for the most efficient folding. Fab fragments present a particular problem because they are unnatural forms of an immunoglobulin and have no evolutionarily designed folding path. Expressing such engineered proteins requires a platform designed to express activities (chaperones) that help them fold in vivo — or have deletions of proteases that might degrade partially folded expressed fragments. Dow has expressed a wide range of antibody derivatives using this technology.
Figure 2 illustrates expression of a Fab fragment with our high-throughput strain-screening technology: 248 unique expression strains were constructed and analyzed by a functionally specific ELISA method. Eight expression plasmids containing three different secretion leaders and arrangements of the Fab-coding genes (transcribed from one promoter) were transformed into 31 different host strains. Those were arrayed in triplicate in deep 96-well plates, then grown up, their genes were induced, the cells harvested and lysed, and ELISA assays were performed. Fab expression was 100–200 mg/L (Figure 2).
Figure 2: ()
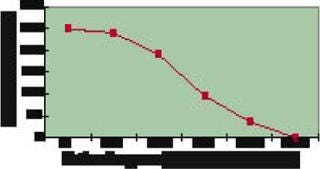
Figure 3 shows that nearly all 248 strains constructed produced a detectable amount of the Fab fragment. However, a smaller subset produced at the highest levels measured, thus reinforcing the high-throughput screening approach to strain selection. These highly productive strains all had host protease deletions, indicating that stability of the expressed fragments may have been a barrier to high-level expression in previously evaluated hosts.
Figure 3: ()
In a second stage of work with the Fab fragment, two of the highest-producing strains were evaluated in parallel small-scale fermentors under various growth conditions. Some conditions tested were variations in growth temperature, pH, induction period, and so on. As Figure 4 shows, some improved expression by 20-fold over that observed in the 96-well experiments, achieving up to 2 g/L expression. Total at-the-bench experiment time (from the start of cloning to confirmation of the 1-L titers) was less than eight weeks.
Figure 4: ()
Expression of a Trimeric Human Cytokine:
The TNF-α human cytokine has been tested in a number of therapeutic indications (6). In addition to its native monomeric form, a synthetic trimeric form may also have value in cancer therapy (7). TNF-α itself contains one disulfide bond, so its trimeric form contains three. The nonnative structure lacks a preprogrammed folding path. Therefore expression and folding to a soluble and active form may depend on host strains engineered to overexpress activities known to benefit such folding. This trimeric form had never been tested appropriately for this application because sufficient quantities were never produced before.
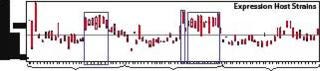
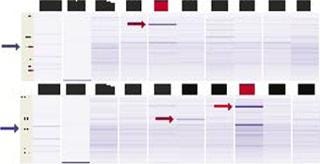
To produce enough material for in vitro and in vivo testing, Dow scientists conducted a 40-strain expression experiment in which four expression plasmids encoding the TNF-α trimer for periplasmic expression were transformed into ten Pfēnex host strains that contained both overexpressed chaperones and protease deletions. As Figure 5 shows, at least two strains (IDs 22 and 32), expressed enough of the trimer to be detected by sodium dodecyl sulfate capillary gel electrophoresis (SDS-CGE, blue arrows, Figure 5) using a LabChip 90 instrument from Caliper Life Sciences (www.caliperls.com). Both strains also overexpress native P. fluorescens chaperones (red arrows, Figure 5). Other strains evidently overexpressing different chaperones (strain ID 30) showed no effect, further validating our parallel high-throughput approach.
Figure 5: ()
Further fermentation range-finding experiments identified a Pfēnex strain producing more than 4 g/L of biologically active trimeric TNF-α (data not shown). In both of these examples, secretion leaders proved to be precisely and quantitatively removed in the strains chosen for further process development.
Current Status of the Platform
The “integrated approach” to downstream processing — in which a target protein’s nature and structure are carefully matched to an expression system — helps ensure overall success at all development stages. In addition to the significant benefits in high-level production of difficult-to-express, nonnative, disulfide-bonded proteins seen herein, periplasmic location of recombinant protein in a Gram-negative bacterium such as P. fluorescens can substantially benefit protein recovery and downstream processing. Roughly 80% of all cellular mass is contained within the cytoplasm of these cells. If the periplasm contents could be released through selective permeabilization of the outer cellular membrane, we could enrich the target protein by fivefold. This would also separate that protein from host-cell contaminants (e.g., DNA). Dow scientists have tested both physical and chemical approaches to periplasmic release at small scale. Promising approaches are confirmed in scalable equipment and, if successful, they decrease the number of unit operations required for purification.
Dow has issued multiple commercial licenses to the Pfēnex platform since 2004, and several more are in negotiation. Current licensees range from large multinational pharmas based in North America, Europe, and India to small, privately held biotechnology companies. Such rapid technology adoption has led to a fast progression in human clinical testing, with our lead licensee completing multiple phase II clinical trials of a protein produced using this technology. Several others expect to file INDs within the next year.
REFERENCES
1.) Schneider, JC. 2005. Auxotrophic Markers pyrF and proC Can Replace Antibiotic Markers on Protein Production Plasmids in High–Cell-Density Pseudomonas fluorescens Fermentation. Biotechnol. Prog. 21:343-248.
2.) Retallack, DM. 2006. Identification of Anthranilate and Benzoate Metabolic Operons of Pseudomonas fluorescens and Functional Characterization of Their Promoter Regions. Microb. Cell Fact. 5:1-10.
3.) Vimberg, V. 2007. Translation Initiation Region Sequence Preferences in Escherichia coli. BMC Mol. Biol. 8:100-112.
4.) Retallack, DM. 2007. Transport of Heterologous Proteins to the Periplasmic Space of Pseudomonas fluorescens Using a Variety of Native Signal Sequences. Biotechnol. Lett. 10:1483-1491.
5.) Chen, C. 2004. High-Level Accumulation of Recombinant Antibody Fragment in the Periplasm of E. coli Requires a Triple Mutant (degP, prc, spr) Host Strain. Biotechnol. Bioeng. 85:463-474.
6.) Wajant, H, J Gerspach, and K. Pfizenmaier. 2005. Tumor Therapeutics By Design: Targeting and Activation of Death Receptors. Cytok. Growth Fact. Rev. 16:55.
7.) Krippner-Heidenreich, A. 2008: in press. Single Chain TNF, a TNF Derivative with Enhanced Stability and Antitumoral Activity. J. Immunol..