Sterilizing-grade filtration is an essential operation for biomanufacturing. It ensures that drug substances are free from microorganisms at the end of a downstream process. The COVID-19 pandemic has highlighted the need for high-quality therapies to be manufactured efficiently at scale, with particular focus on the need for multiple vaccines to be developed, produced, and distributed globally (1). Some vaccines have used lipid nanoparticle encapsulation technology, which also has potential for use in gene therapy development in the near future. Lipid delivery vehicles offer several therapeutic advantages that have contributed to the success of vaccine manufacturing worldwide.
However, lipid nanoparticles can present some difficulties during bioprocessing at the sterilizing-grade filtration step (2, 3). Lipid nanoparticles such as liposomes often are much larger than conventional biologics such as monoclonal antibodies (MAbs). That can be problematic when a particle’s size is reasonably close to the nominal pore size of sterilizing-grade membranes, which is typically 200 nm. Lipid nanoparticles are shear sensitive (and thus could deform during filtration), which could explain unconventional filtration and fouling behavior. Improved understanding of sterilizing-grade filtration for lipid nanoparticles such as liposomes can increase processing efficiency and quality.
Over the past decade, high-resolution, three-dimensional (3D) imaging approaches have been applied extensively for visualizing and characterizing a vast range of complex structures. Two techniques are relevant to the research presented herein: X-ray computed tomography (CT) and confocal laser-scanning microscopy. X-ray CT has been used in other studies to evaluate chromatography structures at multiple length scales. It has the resolution required to represent the internal, detailed geometry of sterilizing-grade membranes. Confocal microscopy is effective for imaging tagged and dyed samples, enabling position-based identification of feed materials. Through those complementing approaches, both filter membrane and liposomes can be visualized and characterized.
Filtration and Imaging
For our study, we used a dual layer polyethersulfone (PES) system for filtration, with the upstream layer being asymmetric and the downstream layer being symmetric with a nominal pore size of 200 nm. Microfluidization produced 200-nm liposomes, with a measured mean diameter of 143 nm and polydispersity index of 0.1, including dyes that were incorporated into liposomes for postprocessing confocal imaging. We filtered those liposomes at constant differential pressures between 0.07 and 0.21 MPa (4).
Researchers at University College London performed 3D imaging using two techniques: X-ray CT (Ultra 810 system from Zeiss) at a 63-nm pixel size (5) for examining membrane structure and confocal microscopy for identifying the location where liposomes and bacteria were retained within those membranes following filtration. Reconstructed 3D files were evaluated and measured using Avizo Fire 9.5 software (Thermo Fisher Scientific). Pore-network maps were generated for producing pore-size–distribution data for each membrane layer (6).
Visualization and Characterization
For X-ray nano-CT imaging, we adhered small membrane pieces to the side of a pinhead, thus demonstrating the small size of samples needed to achieve suitable resolutions. Following imaging, we reconstructed two-dimensional (2D) radiographs into 3D digital volumes, which then could be processed for analysis. From the void phase, a network map could be generated, thus enabling visualization of pores and their interconnectivity for both membrane layers (Figure 1).

Figure 1: 3D renders of sterilizing-grade membranes subvolumes from X-ray computed tomography (CT) imaging; (A) asymmetric membrane material phase; (B) asymmetric membrane pore network; (C) symmetric membrane material phase; (D) symmetric membrane material phase (used with permission from reference 6).
Both the 3D material render and the pore-network map show the membranes’ relative asymmetry and symmetry. Structure is related directly to function and performance, with the asymmetric material used upstream of a symmetric membrane to capture large particles that can foul or block the tighter downstream layer (7). For complex feed streams, assessing fouling accumulation during filtration or over the lifetime of a filter is not straightforward for lipid nanoparticle systems. However, biomanufacturers of high-value products should understand and optimize their processes.
Generating pore-network maps that enable porosity and pore-size measurement helps to increase understanding of complex, internal geometry of bioseparation structures. Such 3D representations can provide the basis for complex flow simulations to evaluate key characteristics such as tortuosity. That parameter has been evaluated using positional analysis to demonstrate the respective asymmetry and symmetry of bioseparation structures (Figure 2).
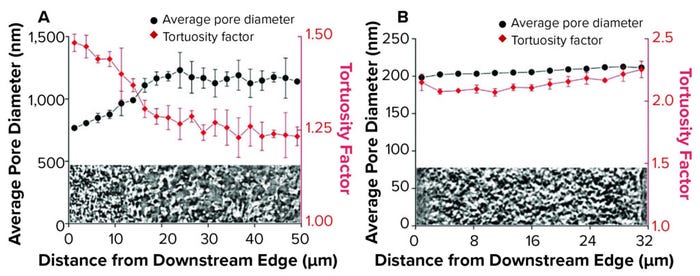
Figure 2: Positional analysis of membrane structure; (A) asymmetric membrane; (B) symmetric membrane.
Although X-ray CT is effective for high-resolution imaging of internal geometry, it is not suitable for imaging samples containing foulant. For that reason, we applied confocal microscopy to view the location of dyed liposomes that had been retained within either membrane layer following filtration (Figure 3).
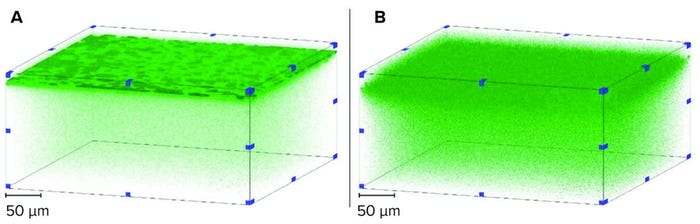
Figure 3: 3D renders of dyed liposomes retained within membrane layers; (A) symmetric
membrane; (B) symmetric membrane; figure originally published as part of reference 6.
In our study, confocal imaging visualized only retained liposomes following filtration, which represented <10% of the total liposomes filtered. In both cases, most liposomes were found in a band between 5 µm and 10 µm into each membrane before reducing throughout the remainder of the sample. In the upstream membrane layer, the liposome-retention peak was particularly concentrated, with 3D reconstructions also displaying intensity disparities at the same distance through the sample. Following research from Folmsbee and Zourna for a differential pressure of interest, we performed filtration under different operating conditions and compared the imaged liposome-retention profiles (3–4) (Figure 4).
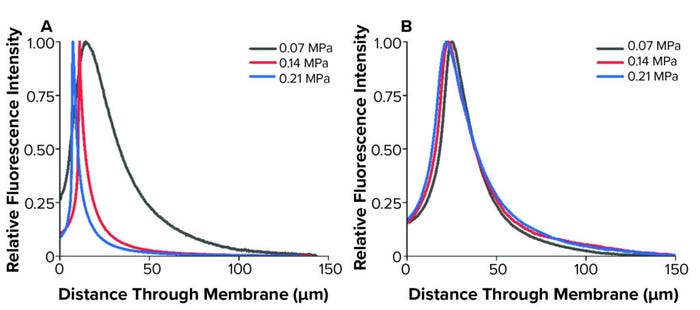
Figure 4: Liposome retention profiles following filtration at multiple differential pressures; (A) asymmetric membrane; (B) symmetric membrane (used with permission from reference 6).
We observed changes to the asymmetric membrane retention profiles because of differential pressure. The symmetric membrane displayed similar profiles at the operational conditions investigated. In the upstream asymmetric membrane, the retention profiles tightened and had peaks closer to the filter surface as differential pressure increased. Zourna has reported that when using this membrane, liposome, and pressure setup, yield and throughput improve substantially as differential pressure increased (4). Such change in filtration behavior under altered operating conditions was apparent with the upstream membrane in our study. However, the downstream membrane had consistent profiles and intensity.
Although those results suggest the need for further investigation, it is generally thought that one mechanism for product loss during this step is liposome absorption to a membrane during filtration. As differential pressure increases, relative residence time will decrease for liposomes passing through filters, thereby decreasing absorption opportunities. The liposomes in this study had an average diameter of 143 nm, measured by dynamic light scattering, with a 45-nm standard deviation.
Although increasing differential pressure appears to be a favorable operating approach, Folmsbee reported an increased risk of bacterial penetration with lipid nanoparticle systems at high pressures (3). Given that the function of sterilizing-grade filtration is to remove bacteria from a process stream, that increased risk is a serious concern. We compared retention profiles between liposomes without bacteria and those deliberately cofiltered with Brevundimonas diminuta. B. diminuta is the standard organism for validating sterilizing-grade membrane filters and can be cultured to produce small cells consistently, presenting a challenge for sterilizing-grade filtration (Figure 5).
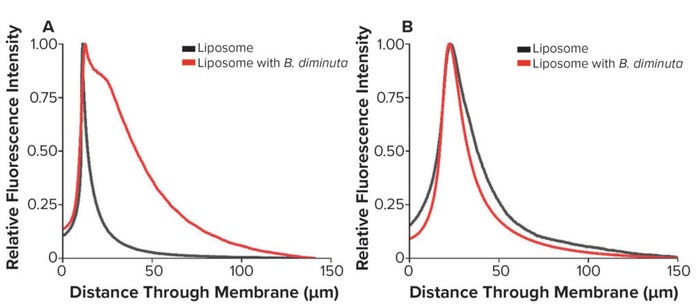
Figure 5: Liposome retention profiles following bacterial cofiltration; (A) asymmetric membrane; (B) symmetric membrane (used with permission from reference 6).
We found that for both membranes, the liposome retention profile was compromised by cofiltration with B. diminuta, indicating interactions between lipid nanoparticles and bacteria that can alter filtration behavior at this step. In the asymmetric membrane, the profile from entrance to peak appears to be similar; however, results show a distinct tail after the peak. In the symmetric membrane, the cofiltered peak occurs in the same position, but the profile is tighter, and the raw intensity value was greater than for the asymmetric membrane.
Zourna found that for this membrane system, increasing differential pressure improved yield and throughput (4). However, Folmsbee raised a concern over bacterial copenetration at high differential pressures, so that is a complex issue for sterilizing-grade filtration (3). Results from Figure 5 show clear differences in bacteria-free and bacteria-mixed feeds. Thus, potential breakthrough must be monitored while process conditions are being optimized. That is particularly important when investigating or optimizing filtrations of new lipid-carrier compositions or size when used with sterilizing-grade membranes.
An Improved Analysis of Filter Membrane Structure
Our study provides insight into the detailed, 3D structures of sterilizing-grade membranes and where liposomes are entrapped within. A greater amount of information can be gathered through implementing two complementary than is possible when using only one approach. Relating imaging results to conventional bioprocessing parameters such as yield and throughput enables a greater understanding of the relationship between membrane structure and performance. Further research on that topic would be beneficial for expanding the number of lipid nanoparticles, membranes, and operational conditions observed so that a robust framework can be developed to correlate results with existing filtration fouling knowledge.
Acknowledgments
The authors thank Nigel Jackson, Stephen Turner, John Welsh at Pall Corporation and Mike Hoare, Francesco Lacoviello, and Paul Shearing at UCL. This research has been part of the UCL–Pall Corporation Centre of Excellence that was established in 2018.
References
1 Sohrabi C, et al. World Health Organization Declares Global Emergency: A Review of the 2019 Novel Coronavirus (COVID-19). Int. J. Surg. 76, 2020: 71–76; https://doi.org/10.1016/j.ijsu.2020.02.034.
2 Watson T. Drug Formulations Are Changing: New Sterile Filtration Challenges in the Changing Landscape of Drug Formulations. BioProcess Int. 18(11–12) 2020: insert; https://bioprocessintl.com/sponsored-content/new-sterile-filtration-challenges-in-the-changing-landscape-of-drug-formulations.
3 Folmsbee M. Evaluation of the Effect of the Volume Throughput and Maximum Flux of Low-Surface-Tension Fluids on Bacterial Penetration of 0.2-Micron–Rated Filters during Process-Specific Filter Validation Testing. PDA J. Pharm. Sci. Technol. 69(2) 2015: 307–316; https://doi.org/10.5731/pdajpst.2015.01026.
4 Zourna K, et al. Optimizing the Filtration of Liposomes Using Sterilizing-Grade Filters. PDA J. Pharm. Sci. Technol. 75(2) 2021: 128–140; https://doi.org/10.5731/pdajpst.2020.011866.
5 Johnson TF, et al. Three Dimensional Characterisation of Chromatography Bead Internal Structure Using X-ray Computed Tomography and Focused Ion Beam Microscopy. J. Chromatogr. A. 1566, 2018: 79–88; https://doi.org/10.1016/j.chroma.2018.06.054.
6 Johnson TF, et al. Liposome Sterile Filtration Characterization via X-ray Computed Tomography and Confocal Microscopy. Membranes 11(11) 2021: 905; https://doi.org/10.3390/membranes11110905.
7 Taylor N, et al. Enhancing the Performance of Sterile Filtration for Viral Vaccines and Model Nanoparticles Using an Appropriate Prefilter. J. Membrane Sci. 647, 2022: 120254; https://doi.org/10.1016/j.memsci.2022.120264.
Further Reading
Eshete DA, Ian Johnson I, Mabery K. The Impact of Protein Stability on Virus Filtration. BioProcess Int. 20(4) 2022; https://bioprocessintl.com/sponsored-content/the-impact-of-protein-stability-on-virus-filtration.
Frei-Rutishauser B, et al. Factors Affecting Sterile Filtration of Sodium-Carboxymethylcellulose–Based Solutions. BioProcess Int. 14(3) 2016; https://bioprocessintl.com/analytical/downstream-development/factors-affecting-sterile-filtration-of-sodium-carboxymethylcellulose-based-solutions.
Mok Y, et al. Best Practices for Critical Sterile Filter Operation: A Case Study. BioProcess Int. 14(5) 2016; https://bioprocessintl.com/downstream-processing/filtration/best-practices-for-critical-sterile-filter-operation-a-case-study.
Watson T. Drug Formulations Are Changing: New Sterile Filtration Challenges in the Changing Landscape of Drug Formulations. BioProcess Int. 18(11–12) 2020; https://bioprocessintl.com/sponsored-content/new-sterile-filtration-challenges-in-the-changing-landscape-of-drug-formulations.
Thomas Johnson is a UKRI research fellow at University College London. Kyle Jones is an R&D manager at Pall Corporation. Kalliopi Zourna is a biotech process R&D manager at Pall Corporation. Corresponding author Daniel Bracewell is a professor of bioprocess analysis at University College London, Gower Street, London, WC1E 6BT; [email protected].










