Advancements in Processing That Optimize Samples for Future Research
Many factors contribute to the quality of biospecimen collections, and most are not mutually exclusive. How we assess the value of a biosample at the time of collection may be very different from at the time of analysis, which can be (and often is) an event in the distant future. To help ensure quality and create a sample resource that is not easily depleted, both novice and experienced “biobankers” can follow some general sample life-cycle management principles to retain the maximum value of their collections.
Sample Collection and Management
Optimized management of research sample assets is best achieved through a comprehensive sample life-cycle management approach. The key stages of this life cycle are planning, collection, transport, bioprocessing, protection, retrieval, and disposal. The most effective process for managing research samples guides each stage with standard operating procedures (SOPs) focused on optimizing the current and future value of each sample asset. By developing an understanding of industry standards and best practices for sample life-cycle management, a research organization can be better positioned to reduce research costs, improve efficiencies, decrease discovery time, and generate more robust and reproducible data for quickly bringing new and improved products to the market.
PRODUCT FOCUS: ALL BIOLOGICALS
PROCESS FOCUS: CLINICAL TESTING
WHO SHOULD READ: PRODUCT DEVELOPMENT, LABORATORY MANAGEMENT, AND ANALYTICAL PERSONNEL
KEYWORDS: ASSAYS, LABORATORY INFORMATION, SAMPLE PREPARATION, DNA AMPLIFICATION, SOPs, BIOBANKING
LEVEL: BASIC
Sample Collection Standards: Most quality biobanks have established protocols for processing samples after collection of clinical or primary specimens. Depending on the associated study, those processes happen either immediately following collection or at some time in the future after the primary sample or some processing intermediate is retrieved from storage. Although some technological variability comes with sample processing as it relates to the ultimate quality (and identity) of a sample, it’s important to establish sample-collection guidelines that ensure long-term sample viability. At the very least, it is important to document each collection because it will directly affect analysis “downstream.”
Literature provides examples of how samples can differ as a function of collection protocols (e.g., tissue biopsies) and the importance of establishing SOPs for sample procurement (1). The same is true for all biospecimens regardless of their tissue source. But not even the best laid plans and protocols can prevent problems that occur during collection that can affect a given sample at any time. Unfortunately, sample collection can never be completely controlled.
That said, documentation of collection-event information is mission critical to qualifying the integrity of samples that will be subject to comparative analysis. Even though specific collection events cannot be controlled, qualification of samples based on accurate and careful documentation can be integrated into the analytical and functional quality control (QC) processes described below. An integrated sample collection, processing, and storage program helps create a set of quality metrics for every sample and all its derivatives. That ensures not only the appropriate use of a given sample, but more important, the interpretation of data derived from analysis.
Essentials for Maximizing Biological Resources
To maximize the potential of every biospecimen, a number of principles can be used to create a high-quality renewable resource. Because of increasing sample collection numbers and requests for access to biologicals, it is imperative that a solid operational plan is created to protect precious research sample resources. The size of most primary clinical samples collected for research is decreasing. There is also a trend toward adopting less invasive sample collection for peripheral biomarker studies (that yield less material than does whole blood as a tissue source). Both suggest the importance of establishing sample collection and processing protocols. An integrated program that ties sample collection to the processing stage of the sample life cycle allows for implementation of important strategies for maximizing preservation, improving extraction efficiency, defining storage formats, and making resources renewable.
Maximal Preservation of Primary Samples: Over the past few years, investigators around the globe have struggled with obtaining high-quality nucleic acids from formalin-fixed paraffin-embedded (FFPE) tissue. This is a prime example of how even the best practices for tissue preservation at one time may not be the best choice for preparing samples for high-quality molecular analyses later. So it is imperative that we continue to use the least amount possible of primary samples for analysis and thus better preserve them for potential applications and new analytical technologies in the future.
An equally important reason for implementing strategies that use minimal amounts of primary samples is to support the trend towards collecting smaller tissue sizes and reducing volumes of liquid biological materials. A growing desire to distribute each sample among more investigators for a wider range of analyses further supports the need for protocols that maximally preserve all primary samples by using less of each sample for standard analyses.
Figure 1 illustrates sample collection and distribution trends for multicenter studies supported by the Rutgers University Cell and DNA Repository (RUCDR). One way to accomplish the goal of maximizing preservation is to better integrate the collection of samples directly with biosample processing laboratory activities so that appropriate strategies are implemented.
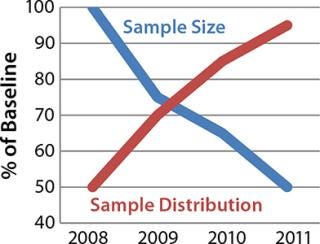
Figure 1: ()
Use of Efficient Extraction Methodologies: With advanced chemistries and automation platforms, biorepositories are better suited to handle an increased volume of samples commensurate with today’s clinical trials and large disease-based studies. However, more tissue sources and biomaterial components require careful selection of extraction methodologies. For example, as the trend for noninvasive DNA collection (e.g., saliva and buccal swabs) continues , we must critically evaluate our methodologies for DNA extraction.
First, we must ensure that captured DNA is of human origin (for most
studies) and that it is suitable for sensitive downstream analysis (next-generation sequencing). Extraction chemistries bundled with collection kits are often not the most efficient choice for sample preparation. Figure 2 compares the results of saliva DNA extractions using a collection-kit manufacturer’s protocol with a bead-based chemistry that provides better yields and quality.
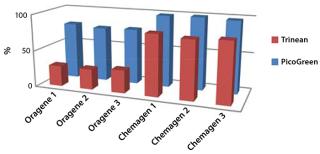
Figure 2: ()
Extraction of RNA from Paxgene RNA blood tubes is another example of maximizing the utility of every sample. Traditionally, miRNA is lost in solid-phase extraction technologies or ends up in the total RNA fraction with an organic extraction. Neither approach efficiently manages the needs of a biorepository to store separate fractions isolated during extraction for downstream sample analysis. The RUCDR established (in collaboration with Qiagen) an automated work flow to store samples enriched for miRNA in a separate fraction from total RNA using an automated platform that now preserves the maximum potential for all RNA components of a single primary sample. Such approaches help ensure the utility of many biosample components for later analysis.
Implementing Defined Storage Formats: An essential component to biospecimen management is the ability to efficiently access, organize, and distribute derivatives of primary samples while preserving optimal environmental storage conditions. Historically, the balance among storage vessels, efficient sample distribution, and economical management of collections was compromised by expensive storage vessels that required manufacturer-specific automated storage infrastructure. In addition, the volumes of primary stock samples were driven by the up-front processing and collection teams who did not take into consideration more efficient storage (and retrieval) scenarios.
An advantage to the complete integration of the sample life cycle is selection of defined storage formats that preserve biomaterials through different environmental conditions. This creates a uniform storage format to maximize precious freezer space and allows a laboratory to automate and rapidly access biospecimens for customized distribution formats compatible with downstream analytical applications. Several manufacturers have developed products that can be integrated into existing bioprocessing workflows without requiring specific storage automation.
For example, the RUCDR partnered with Micronic Inc. to implement a line of laser-etched, two-dimensional (2D) barcoded tubes designed specifically for biorepository applications. The tubes are organized in racks that take up 35% less freezer space than conventional 2-mL tube boxes. They are also completely compatible with all Society for Biomolecular Sciences (SBS) automation standard for aliquoting and tube handling. These tubes can be sealed with either pierceable septa or screw caps, using gaskets for biomaterials that require additional protection from the environment. Handling is easy with inexpensive stand-alone automation, or the tubes can be integrated into several automated storage environments, making sample organization and retrieval for distribution (or analysis) an exacting, efficient process. Because storage vessels are integral to so many steps in the sample life cycle, it is imperative that the processing and storage components of every project are integrated to take advantage of operational and storage efficiencies now available across biospecimen tissue sources.
Creating Renewable Resources Through Cellular and Molecular Approaches: Even with the best-laid plans and a fully integrated biosample processing and storage program, some samples will ultimately become a limiting resource—either due to the starting amount of primary material or the global demand of a specific set of samples or their molecular derivatives. To create a truly renewable resource from limited amounts of materials a bioprocessing program needs to be able to support both cellular and molecular techniques that yield a product that can be qualified for downstream analyses.
Two approaches used at the RUCDR are seamlessly integrated with both storage and sample management operations to manage the amount of biomaterial that remains in a given collection and tracks its use through biomaterial requests. The first approach (used for most whole-blood samples) is establishment of cryopreserved lymphocytes (CPLs) and/or lymphoblastoid cell lines (LCLs). This process creates a truly renewable resource for DNA that has been validated for genotyping and copy-number–variant (CNV) applications. It also provides a resource for high-throughput screening applications in drug discovery and development. An added advantage of CPLs is the further ability to create induced pluripotent cell lines (iPCs) for developmental studies. Creation of any cell line from a primary sample creates opportunities for derivatives that otherwise could become a limiting resource.
A different approach is to create a renewable resource through nucleic-acid amplification. Both DNA and RNA can be amplified using a minimal amount of starting material, which ultimately yields enough product for both analysis and sample archival. An enabler of this strategy is the integration of NuGEN’s Ovation technology into the RNA extraction and archival process at the RUCDR. Given the lability of RNA and the need to limit sample manipulation and freeze–thaw cycles, that methodology linearly amplifies cDNA off an RNA template (500 pg) and creates a technology- and platform-agnostic product that could be used for quantitative polymerase chain reaction (QPCR), microarrays, and RNA sequencing across multiple laboratories. Yields of cDNA from this process also provide material that can be archived for future analyses. Given the small amount of RNA needed for amplification, this strategy maximizes preservation of the primary RNA sample. Operational efficiencies aside, the main advantage of an approach like this is development of a robust biomaterial that can be compared more robustly across sites performing gene expression analysis.
Figure 3(LEFT) compares the stability of an amplified cDNA product with total RNA demonstrating improved fidelity of cDNA as a template for gene-expression analysis. Figure 3(RIGHT) illustrates improved laboratory-to-laboratory performance achievable using amplified cDNA (rather than RNA) supplied to individual laboratories. Like some other approaches, this approach requires a seamless connection among all components of a sample’s life cycle, which in turn has synergistic effects, not only on sample preservation, but also performance.
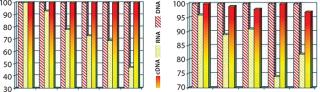
Figure 3: ()
>
Integrating Analytical and Functional QC
The most important component of a sample-processing work flow is inclusion of biosample quality control. Traditionally, QC has been limited to analytical analysis, which qualifies the static or “biostorage” component of quality assessment. A number of technologies are used for making analytical measurements that provide data to quantify and qualify samples based on metrics that are surrogates of sample quality. For instance, blood plasma is often qualified based on its color and opacity measurements as a function of sample quality. By contrast DNA is quantified through spectroscopy (or other quantification methods) to measure its concentration and purity through absorbance ratios that act as surrogates for a global sample quality measurement.
Although such measurements are critical for quantitation of some biomaterials, it is becoming important to functionally qualify samples by interrogating sample identity, contamination, and performance for downstream analyses. To that end, a number of analytical approaches are being used to provide a more comprehensive assessment of sample quality (and annotation) at the time of processing — and in some instances to measure performance as a function of time in storage. Two such approaches are the RUID sample qualification panel for DNA-based derivatives and the RUIDGx RNA fidelity tool for qualifying mRNA transcripts in gene-expression analysis. Both technologies developed at the RUCDR help complete the QC process for nucleic acids extracted regardless of tissue source.
Briefly, the RUID sample qualification panel consists of 96 single-nucleotide polymorphisms (SNPs) that are highly polymorphic and have been strategically selected to predict DNA integrity as a function of specific and nonspecific degradation events, thereby correlating SNP call rates with the performance of a given sample in a range of downstream analytical analyses. The panel is also designed to determine gender, ethnicity, and parentage, thereby providing useful nonpersonal health-identifying information that can be used in quality assurance of a sample collection and annotation process. This quick, high-throughput, and cost-effective strategy provides an instant assessment of sample uniqueness (the 96-SNP profile is unique for every 2 × 1024 individuals) and potential sample contamination.
Samples being used to create renewable resources (through cell-line creation or nucleic-acid amplification) can be reconciled and, more important, validated across complex collection schedules and/or laboratory procedures. Similarly, an approach has also been created and implemented for measuring cDNA fidelity as a function of gene-expression analysis. That approach uses standard QPCR chemistries to measure transcript abundance on a tissue-specific level, taking into consideration both expression levels and the temporal integrity of tissue specific transcripts.
Sample-Storage Standards
Appropriate sample-storage practices are critical to maintaining the integrity of biological samples. Unfortunately, such practices can vary greatly among different institutions and laboratories. Standards are commonly established based on current facilities to meet the desired needs of specific analytical techniques. That does not necessarily meet the requirements of a global sample management infrastructure (2). Organizations such as the National Cancer Institute (NCI) and International Society for Biological and Environmental Repositories (ISBER) publish best practices and continually make advancements as sample-management technologies change (1, 3). In addition, the College of American Pathologists has initiated a biorepository accreditation program that will begin to unify some of those practices. BioStorage Technologies, Inc. — a global comprehensive sample management solutions provider — has published good-storage–practice guidelines that are used by many companies and government organizations as a foundation for developing best-in-class storage practices to support their research.
Long- or Short-Term Storage: Regardless of the duration of storage, consistency is key to ensuring the molecular integrity of a specimen. Here are some important components to consider for maintaining reliable and dependable sample-storage conditions:
Properly maintained freezers and liquid nitrogen (LN2) dewers, including records and logs of maintenance activities
Secure facilities capable of holding equipment required for sample processing and storage
Trained staff
Sample inventory technology capable of managing complex and unique collections at varying levels of specificity
Reserves of all samples stored in multiple aliquots and separate freezers
Thorough and detailed, regulatory-compliant SOPs and audit-trail sample tracking
Regulatory-compliant certification of facilities and systems
Constant temperature monitoring
Redundant processes, equipment, and facilities for emergency situations and business continuity.
SOP Documentation
Even the best practices are ineffective if they are not followed. Implementation of a standardized practice begins with developing a document management system that includes SOPs and training files. Newly implemented SOPs should be reviewed by several individuals, with a quality-assurance representative as the final approver. Laboratory personnel should be trained on each SOP before they execute a given activity. And SOPs should be reviewed annually to ensure that they are up to date and properly represent the processes being performed. Routine audits can ensure that SOPs are being followed appropriately.
Centralization and Auditing
Given the large number of patients enrolled in clinical studies, it is easy to understand why the resulting clinical samples get scattered all over the globe (4). This situation often requires additional effort, time, and resources in locating specific samples. We recommend a centralized sample-management approach for faster access to sample inventories and reduced timelines for sample collection and transportation to research laboratories. If a centralized approach is impossible, then an information technology (IT) infrastructure needs to unify separate facilities in their sample inventory management and reporting.
Audit Trails and Data Management: Electronic audit trails verify the location and movement of samples and ensure that those samples have been stored in optimal conditions. Such information needs to follow a sample throughout its life cycle, so a comprehensive data management system is preferred. Factors such as temperature, location, handling, and shipping conditions should be recorded and maintained for the life of a sample.
A Strategic Alliance
RUCDR Infinite Biologics, a global leader in the processing and biobanking of samples for genetic, gene, and cell-based research; and BioStorage Technologies, Inc., the leading global provider of comprehensive sample management solutions, have launched a valuable strategic alliance that provides to the bioscience industry an integrated, state-of-the-art scientific approach and technology infrastructure for delivery of advanced sample bioprocessing and biobanking solutions.
Additional data elements that could be part of a sample-management technology system include study identifiers, patient-consent tracking, and sample-quality coding (e.g., purity, yield and concentration obtained following sample processing procedures). Such information should be maintained and linked to each sample for as long as required. Management of this level of data should be securely controlled according to US FDA 21 CFR Part 11 requirements. Other regulations to consider are the Health Insurance P
ortability and Accountability Act (HIPAA) in the United States and the Data Protection Act in the United Kingdom.
Good Sample Management Supports Bioprocessing
Biopharmaceutical companies that develop robust, regulatory-compliant, and comprehensive sample management technology systems are empowering their research teams. Consolidated sample inventory data can be combined with clinical result databases to support improved sample selection for future research studies. Faster selection of higher quality research samples enables companies to shorten their research timelines and to bring improved therapies to the market at substantially lowered costs. This process benefits research organizations, patients, health-care providers, and payers.
About the Author
Author Details
Dr. Andrew Brooks is chief operating officer of Rutgers University Cell and DNA Repository (RUCDR) Infinite Biologics, 604 Allison Road, Piscataway, NJ 08854; 1-848-445-0225, fax 1-732-445-0152; [email protected], www.rucdr.org. Aleks Davis is associate director of comprehensive solutions and scientific advisor at BioStorage Technologies, Inc., 2910 Fortune Circle West, Suite E, Indianapolis, IN 46241; 1-317-268-5500, www.biostorage.org. RUID and RUIDGx are registered trademarkes of the RUCDR.
REFERENCES
1.) Office of Biorepositories and Biospecimen Research NCI Best Practices for Biospecimen Resources, National Cancer Institute, Bethesda.
2.) Olson, S, and A. Berger. 2011.Establishing Precompetitive Collaborations to Stimulate Genomics-Driven Product Development: Workshop Summary, Institute of Medicine (US) Roundtable on Translating Genomic-Based Research for Health, National Academies Press (US), Washington.
3.) 2012. International Society for Biological and Environmental Repositories. 2012 Best Practices for Repositories. Biopreserv. Biobanking 10:79-161.
4.) Elliott, P, and T. Peakman. 2008. The UK Biobank Sample Handling and Storage Protocol for the Collection, Processing, and Archiving of Human Blood and Urine. Int. J. Epidemiol. 37:234-244.
You May Also Like





