Improving IEX Throughput and Performance with Differentiated Chromatography Sorbents
Optimized upstream processing and high-productivity cell culture increase not only target protein titers, but also impurity and contaminant concentrations to be removed from large volumes of feedstock. Simultaneously, biopharmaceutical drug production is increasingly driven by manufacturing cost reduction. These facts together increase the pressure on downstream processing and create an urgent need for more productive and streamlined chromatography operations. Key parameters to consider for enhanced process economics in chromatography are higher protein binding capacities at high flow rates (to reduce batch processing duration) and improved sorbent selectivities (to reduce the number of column chromatography steps and decrease purification cost).
Ion-exchange (IEX) chromatography is one of the most broadly used techniques for protein purification. Protein binding during ion exchange is driven by a combination of factors including primarily
characteristics of the functional group and its density at the surface of the base matrix
pH and conductivity of the mobile phase that drives electrostatic interactions between proteins and functional groups
the nature of the base matrix.
PRODUCT FOCUS: PROTEINS
PROCESS FOCUS: DOWNSTREAM PROCESSING
WHO SHOULD READ: PROCESS DEVELOPMENT, ANALYTICAL
KEYWORDS: ANION-EXCHANGE, CATION-EXCHANGE, PH, GRADIENT ELUTION, SELECTIVITY, DYNAMIC BINDING CAPA#CITY, CONDUCTIVITY
LEVEL: INTERMEDIATE

Many developments occurred in the past decade to enable manufacture of new mechanically stable base matrices and more capacitive IEX sorbents. This progress resulted mainly in enhanced capacities at high process flow rates or shorter residence times (RTs), but resin selectivity has not been substantially modified or usually taken into account. In the past few years, scientists have tried to characterize more precisely the factors that affect the selectivity of various IEX chromatography sorbents.
Wu and Walters showed that different ligand densities could alter the selectivity and order of protein elution for different ion exchangers (1). Harinarayan et al. demonstrated the existence of an exclusion mechanism on ion exchangers that leads to an optimal dynamic binding capacity (DBC) for intermediate pH and conductivity conditions even if optimum conditions for protein binding are expected to be at low conductivity and around the pH value at which the protein is more charged (2). Additionally, Hardin et al. measured that resins with large pore sizes — or smaller pores but low ligand density — could reduce the impact of the exclusion mechanism (3). Altogether, these data suggest that the selectivity of a given ion exchanger is ruled by a complex combination of parameters.
The new Q and S HyperCel IEX chromatography sorbents from Pall Life Sciences are designed to provide a very high protein binding capacity even at low RT, along with a new selectivity and a differentiated salt sensitivity. Low to moderate density of quaternary amine (Q, 99–138 µeq/mL) and sulfonic acid (S, 59–84 µeq/mL) groups are immobilized on a robust and scalable cross-linked cellulose HyperCel matrix that confers low nonspecific binding with chemical stability and excellent mechanical properties.
Here, we characterized the differentiated selectivity of both those sorbents. We first evaluated the DBC for model proteins in various pH, conductivity, and RT conditions compared with other commercially available sorbents: rigid agarose Q and S and polymeric Q and S. We also separated a mixture of model proteins to further characterize the specific selectivity of Q and S HyperCel sorbents. Finally, we performed a real-case application using Q HyperCel sorbent as a capture step for purification of a recombinant green fluorescent protein (rGFP) from an Escherichia coli lysate, and we compared that performance to two other anion exchangers: rigid agarose Q and Q Ceramic HyperD F sorbents.
Materials and Methods
Chemicals and Equipment Used: Analytical-grade reagents came from Sigma Aldrich (www.sigmaaldrich.com) except for bovine serum albumin (BSA), which was provided by Millipore (www. millipore.com). Professor Xavier Santarelli of Ecole Nationale Supérieure de Technologie des Biomolécules de Bordeaux (ENSTBB) in France kindly provided clarified E. coli lysate containing recombinant greenfluorescent protein (rGFP).
The rigid S agaroseorbents came from GE Healthcare (www.gehealthcare. com). Polymeric sorbents came from Tosoh Bioscience (www.tosohbioscience.com). All HyperCel and HyperD sorbents came from Pall Life Sciences (www.pall. com). Each sorbent was packed according to manufacturer instructions into 0.5-cm diameter columns (1 mL with 5.0-cm bed height) from Kronlab (www.ymc-europe.com/ymceurope/ products/preparativeLC/LC_ IntroForKronlab.html). We used an ÄKTAexplorer 100 system from GE Healthcare (www.gelifesciences.com) for all chromatographic runs.
For sodium-dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), we used NuPAGE 4–12% Bis-Tris gels from Invitrogen (www. invitrogen.com) stained with Coomassie SimplyBlue (also from Invitrogen). Total protein content was measured with a bicinchoninic acid (BCA) protein assay from Thermo Scientific Pierce (www. piercenet.com).
Comparison of Anion-Exchange Sorbents Used As a Capture Step for rGFP Purification from E. Coli Lysate: We screened different anion exchangers (Q HyperCel, rigid agarose Q, and Q Ceramic HyperD F) for their selectivity using linear salt-gradient elution. Before equilibration in 50-mM Tris-HCl at pH 8.0, a 100-µL sample of crude E. coli lysate was loaded. After an equilibration buffer wash, elution was performed using salt gradient over 20 column volumes (CV) using 50 mM Tris-HCl at pH 8.0 with 0.5 M NaCl before a final strip with 50 mM Tris-HCl at pH 8.0 with 1.0 M NaCl. The flow rate applied was 1 mL/min.
We transferred the initial salt gradient elution to a four-step elution according to the conductivity at rGFP elution. The sorbent was equilibrated with 50 mM Tris-HCl at pH 8.0, then an 8-mL sample of E. coli lysate was loaded onto the column at 0.5 mL/min (2 min RT). After an equilibration buffer wash, elution was carried out in steps using 50 mM Tris-HCl at pH 8.0 with 0.1, 0.2, 0.5, and 1.0 M NaCl.
We evaluated the DBC for GFP of the sorbents tested at 10% breakthrough by loading E. coli lysate onto the columns until complete saturation. Samples were diluted tenfold in equilibration buffer to delay GFP breakthrough so that saturation was clearly observed. We noted the maximum absorbance value and calculated DBC with the volume loaded until flow-through absorbance was at least 10% of this maximum.
Results and Discussion
Salt/Charge Sensitivity — Influence of pH, Conductivity, and RT on DBC Using Design of Experiments (DoE): We used a design of experiments (DoE) with a central composite face-centered model (on MODDE software from Umetrics AB, www.umetrics.com) to explore the influence of various pH values, conductivities, and RTs on the DBC for model proteins. We chose BSA for anion exchangers and polyclonal human IgG for cation exchangers. Table 1 lists the conditions we tested.
Table 1: Experimental plan for studying the effects of pH, conductivity, and residence time (RT) on dynamic binding capacities* of Q and S HyperCel sorbents, rigid Q and S agarose sorbents, and polymeric Q and S sorbents
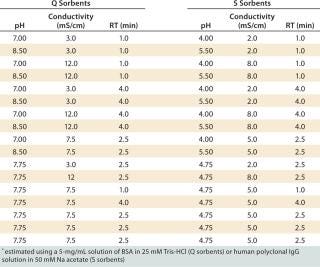
Table 1: Experimental plan for studying the effects of pH, conductivity, and residence time (RT) on dynamic binding capacities* of Q and S HyperCel sorbents, rigid Q and S agarose sorbents, and polymeric Q and S sorbents ()
Contour plots for Q and S HyperCel sorbents (Figures 1 and 2) show that RT does not affect the DBC within the studied range of one to four minutes. The situation is different for the two other sorbents we tested: RT clearly influences the DBC for rigid S agarose (Figure 2, MIDDLE) and polymeric S (Figure 2, RIGHT) sorbents, but not for rigid agarose Q (Figure 1, MIDDLE) and polymeric Q (Figure 1, RIGHT) sorbents. So diffusion did not limit DBC for Q and S HyperCel sorbents, although such a limitation seems to exist for both rigid agarose and polymeric sorbents. The diffusion limit seems to most influence large molecules (as observed for IgG on rigid S agarose and polymeric S).
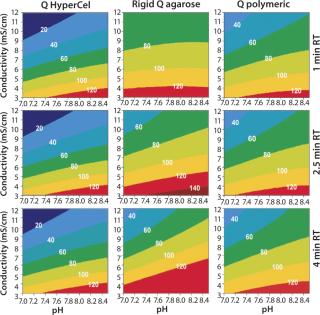
Figure 1: ()
Those contour plots also show that optimal conditions for achieving high DBC on Q and S HyperCel sorbents are found at low conductivity and pH values at which the protein is more charged. This favors the exclusive involvement of ionic interactions between sorbents and proteins, probably with no exclusion mechanism impact. This “pure ion-exchange behavior” should give more predictable separations based on protein isoelectric point (pI) values.
Contour plots achieved for rigid S agarose (Figure 2, MIDDLE) and polymeric S sorbents (Figure 2, RIGHT) are clearly very different from those obtained using S HyperCel sorbent. The DBC decreases with extremes of pH and conductivity, whereas an optimal DBC is achieved at intermediate pH (4.6–5.2) and moderate conductivity (>6 mS/cm). This specific salt/charge behavior illustrates the involvement of other than pure ionic interactions and the possible presence of factors disturbing the charge interactions, such as the exclusion mechanism described by Harinarayan et al. (2). For rigid agarose Q and polymeric Q sorbents, the exclusion mechanism apparently does not play a strong role. As for Q and S HyperCel sorbents, optimal DBC comes under optimal pH and conductivity conditions.
As suggested by Hardin et al. (3), the exclusion mechanism is enhanced for sorbents that present a high ligand density. So the low ligand density of Q and S HyperCel sorbents could possibly explain the mechanism’s absence. For rigid agarose and polymeric sorbents, the ligand density is probably higher and thus leaves room for it.
Our data show differentiated salt/charge behavior for Q and S HyperCel sorbents, probably attributable to their specific base-bead design. We observed different results of protein adsorption on HyperCel sorbents from those on polymeric and rigid agarose ion exchangers. This is in accordance with the work of Hardin et al. (3) and Hubbuch et al. (4), which involved confocal laser scanning microscopy and showed that proteins may have different diffusion mechanisms depending on the solid-phase construction and chemical environment (pH, ionic strength). The pure IEX behavior unaffected by exclusion mechanisms or other interactions we observed for HyperCel sorbents should lead to more predictable separations based on protein pI values. Such a difference in salt/charge sensitivity could also affect the selectivity of these sorbents.
Selectivity — Separation of a Model Protein Mixture with a Linear Salt Gradient: We performed a selectivity test using a mixture of model proteins to study the differentiated salt/charge sensitivity impact. Conditions were exactly the same for all sorbents (Table 2). We used the following proteins: cytochrome C (2 mg/mL, pI 10.2), transferrin (10 mg/mL, pI 6.7), and BSA (10 mg/mL, pI 4.7) in 50 mM Tris-HCl at pH 8.5 for anion exchangers; and lysozyme (5 mg/mL, pI 11.3), cytochrome C (2.5 mg/mL), and ovalbumin (10 mg/mL, pI 4.6) in 50 mM sodium acetate at pH 4.5 for cation exchangers. Figure 3 presents the resulting chromatograms.
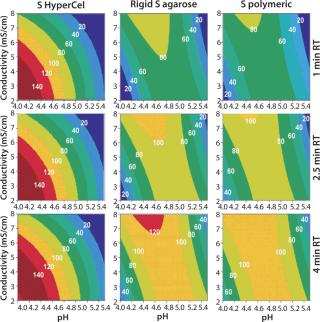
Figure 2: ()
Table 2: Conditions used for separation of a model protein mixture with a linear salt gradient on Q/S HyperCel, rigid Q/S agarose, and Q/S polymeric sorbents

Table 2: Conditions used for separation of a model protein mixture with a linear salt gradient on Q/S HyperCel, rigid Q/S agarose, and Q/S polymeric sorbents ()
Cytochrome C (first peak) was not retained on any anion-exchange sorbent, and it eluted before the gradient started. This is fully expected because cytochrome C is a basic protein that’s positively charged at pH 8.5. Separation of transferrin and BSA along a salt gradient was effective on all sorbents for both proteins and followed the expected elution order based on their pI values. The less acidic protein (transferrin) eluted first, followed by the more acidic BSA. When comparing the elution profiles for different sorbents, we observed that transferrin and BSA eluted earlier from Q HyperCel sorbent than from rigid agarose Q and polymeric Q sorbents, for which the elution conductivity is more or less the same.
As for cation exchangers, differences among the S sorbents were even more pronounced. All three loaded proteins are bound, but only the S HyperCel sorbent could separate them all along the salt gradient. As an acidic protein, ovalbumin is expected — if bound — to elute at the very beginning of the salt gradient; this is actually the case for S HyperCel sorbent but not for rigid S agarose and polymeric S sorbents. Even though both are basic proteins, the clear pI difference between cytochrome C (pI 10.2) and lysozyme (pI 11.3) should allow separation along the salt gradient; this too is the case on the HyperCel sorbent but not on the others.
Our data show that the differentiated salt/charge behavior of Q and S HyperCel sorbents observed during the DoE study affects selectivity among different proteins in a mixture. This observation is critical when considering the use of IEX sorbents for purification of recombinant proteins from real feedstocks.
Capture of a Real Feedstock — Comparing Anion Exchange Sorbents Used As a Capture Step for rGFP: We performed a real-feedstock separation using a clarified E. coli lysate expressing rGFP (absorbance at 490 nm). We selected Q HyperCel sorbent and compared it with rigid agarose Q and Q Ceramic HyperD F anion exchangers.
The first separation used a linear salt gradient from 0 to 0.5 M NaCl to evaluate the selectivity of those three anion exchangers. Figure 4(TOP) shows the chromatograms, with the rGFP elution peak indicated by a red arrow. Absorbance at 280 nm (blue trace) clearly indicates a distinct selectivity for each sorbent. Five or six major peaks were detected in each case, but none of the chromatography profiles were identical. Some similarities could just be seen between the Q Ceramic HyperD F and HyperCel sorbents. Absorbance at 490 nm (red trace) also supported that different general selectivity while additionally highlighting a differentiated salt sensitivity because the rGFP peak eluted at a different conductivity for each sorbent. Interestingly, rGFP eluted from Q HyperCel sorbent at only 13.5 mS/ cm, whereas the other sorbents required higher salt concentration for its elution (19 mS/cm for rigid agarose Q, 16.5 mS/ cm for Q Ceramic HyperD F sorbent). We confirmed the presence of rGFP in elution fractions with SDS-PAGE analysis (data not shown). Several contaminants were also present, and no clear difference in purification effect was seen.
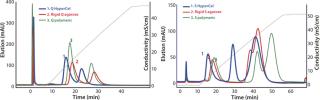
Figure 3: ()
Based on the results of our preliminary gradient elution screening, a step elution of four steps at 0.1, 0.2, 0.5 and 1.0 M NaCl was achieved. The elution profiles showed clear differences (Figure 4, BOTTOM). For rigid agarose Q and Q Ceramic HyperD F sorbents, rGFP was detected at 490 nm in the first elution step (0.1 M NaCl) but mainly eluted during the second step (0.2 M NaCl). For Q HyperCel sorbent, rGFP eluted exclusively during the first step and at 490 nm; no peak was detected in the other elution fractions. Those results confirmed previous data showing the distinct selectivity of Q HyperCel sorbent.
SDS-PAGE analysis of the fractions (Figure 5) was consistent with the absorbance signal at 490 nm recorded during the chromatographic runs. With the rigid agarose Q and Q Ceramic HyperD F, rGFP was detected in the elution fraction at 0.1 M NaCl (lanes E1 and E9), but it mainly eluted at 0.2 M NaCl (lanes E2 and E10). With Q HyperCel sorbent, it was exclusively detected in elution fraction at 0.1 M NaCl (lane E5). So a single elution step at 0.1 M NaCl was sufficient to collect all the rGFP bound on Q HyperCel sorbent, whereas 0.2 M NaCl was required to completely recover the rGFP from Q Ceramic HyperD F and rigid agarose Q sorbents. The large amount of contaminants eluting at 0.2 M NaCl were not present at 0.1 M NaCl, which implies that rGFP recovered from the Q HyperCel column is the most pure. So it would greatly facilitate further orthogonal purification steps and enhance the efficiency of an entire downstream process.
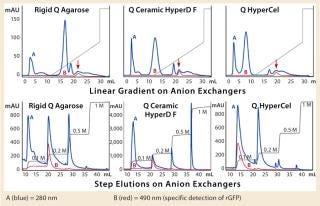
Figure 4: ()
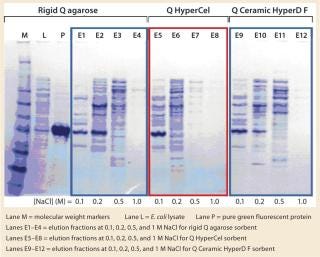
Figure 5: ()
We evaluated DBC of the media for rGFP with breakthrough (BT) curves following the absorbance signal at 490 nm. The E. coli lysate was loaded onto 1-mL columns packed with the different sorbents until saturation was reached. We took the saturation plateau to calculate the 10% BT absorbance level and then calculated DBC at 10% BT (Table 3). DBC results showed significant differences: 65% higher for Q HyperCel sorbent than for the rigid agarose Q sorbent. That higher DBC is probably correlated to the different selectivity and salt sensitivity. The population of bound or unbound molecules is likely to be very different on rigid agarose Q and on Q HyperCel sorbent. We assume that the rigid agarose Q sorbent is capturing more contaminants than is the Q HyperCel sorbent in these conditions.
Table 3: Dynamic binding capacity for rGFP on Q Ceramic HyperD F, Q HyperCel, and rigid Q agarose media; DBC was estimated at 10% breakthrough after column saturation for all sorbents

Table 3: Dynamic binding capacity for rGFP on Q Ceramic HyperD F, Q HyperCel, and rigid Q agarose media; DBC was estimated at 10% breakthrough after column saturation for all sorbents ()
All data obtained for the capture step of rGFP purification show that the specific selectivity and salt/charge behavior of Q HyperCel sorbent affects the separation of the target protein from contaminants. At full manufacturing scale, these differences may have a positive impact on global process economics and lead to better productivity.
Selecting for Selectivity
We’ve explored how a new selectivity can be invented for ion exchange based on the specific design of chromatography sorbent beads. Even carrying in principle the same functional cation-or anion-exchange chemical groups, sorbents with different “base-bead” designs have completely differentiated protein selectivities. High binding capacities for proteins and good mechanical and flow rate properties remain important factors at manufacturing scale. Moreover, we suggest that selectivity (which is often neglected) should be also carefully considered to improve process economics. In that respect, modern IEX matrices such as Q and S HyperCel sorbents may provide efficient and differentiated separation solutions and should be included in early resin screening procedures. The assumption that their behavior is ruled by a pure ion-exchange mechanism that favors a more predictable separation.
About the Author
Author Details
Sylvio Bengio, PhD, is scientific communication manager; Stéphanie Chaudoreille, PhD, and Jérôme Champagne, PhD, are applications R&D scientists; and corresponding author René Gantier, PhD, is applications R&D manager for Pall BioSepra, a division of Pall France, 48 avenue des Genottes, 95800 Cergy Saint Christophe, France; 33-1-3420-7835, fax 33-1-3420-7878; [email protected]; www.pall.com.HyperD and HyperCel are trademarks of Pall Corporation. MODDE is a trademark of MKS Umetrics AB. ÄKTAexplorer is a trademark of GE Healthcare. NuPage is a trademark of Invitrogen.
REFERENCES
1.) Wu, D, and RW Rodney. 1992. Effects of Stationary Phase Ligand Density on High-Performance Ion-Exchange Chromatography of Proteins. J. Chromatogr. 508:7.
2.) Harinarayan, C. 2006. An Exclusive Mechanism in Ion-Exchange Chromatography. Biotechnol. Bioeng. 95:775.
3.) Hardin, AM. 2009. Ion-Exchange Chromatography of Monoclonal Antibodies: Effect of Resin Ligand Density on Dynamic Binding Capaicty. J. Chromatogr. A 1216:4366.
4.) Hubbuch, J. 2003. Mechanism and Kinetics of Protein Transport in Chromatographic Media Studied By Confocal Laser Scanning Microscopy, Part II: Impact on Chromatographic Separations. J Chromatogr. A 1021:105.
You May Also Like





