Control of Host Cell Proteins in Monoclonal-Antibody Bioprocessing: Using Proteomic Analysis To Understand Impurity Clearance and Persistence During Purification
Downstream process development can proceed like a detective novel, starting with evidence of something seriously wrong and rapidly evolving into a “whodunit.” The evidence often comes as precipitate particles in what is supposed to be a stable formulation. The whodunit takes the form of root-cause analysis into the degradation mechanism of a biopharmaceutical product or of a key ingredient in its formulation. And the culprit often turns out to be an enzyme present in such small quantities as to be almost undetectable. The rash of such cases during manufacturing of monoclonal antibodies (MAbs) and other biopharmaceuticals has changed the field of impurity clearance, including associated assays and control strategies.
Issues with impurity clearance stem almost invariably from host cell proteins (HCPs). Along with host-cell DNA (hcDNA), cell debris, lipids, and viruses, HCPs are categorized as process-related impurities, distinguishing them from product-related impurities such as product aggregates (often referred to as species of high molecular weight (HMW)) and fragments. Historically, the extremely large and heterogeneous class of HCPs has been considered as a unit, both for specific analyses (e.g., as a set of impurities to be detected using enzyme-linked immunosorbent assays (ELISAs)) and for assessment of downstream clearance, with overall levels in a MAb drug substance needing to fall below the target of 100 ppm (ng HCP/mg MAb) and with products typically having ~10 ppm. Cases in which low concentrations of individual HCPs were found to have deleterious effects on drug substances, formulations, and recipients have since led to classification of dozens of specific species as “difficult-to-remove” or “high-risk” (1). The latter category includes various proteases such as cathepsins, lipases, and hydrolases.
Identification of a single problematic HCP can prompt downstream scientists to give it specific analytical attention and make focused efforts to ensure its removal to sufficiently low levels, preferably below a method’s limit of detection. Cases of such adaptation have directly influenced current approaches to process development, including considerations for associated analytical support. For instance, although platform processes for MAb purification have been well established for about two decades (2, 3), and despite the emergence of liquid chromatography with mass spectrometry (LC-MS) as an enabling technology for HCP analysis during process development and (increasingly) manufacturing, proteomic analyses of MAb process streams are shedding light on new biophysical aspects of downstream processes. Here, we overview that rapidly evolving field, specifically for proteomic assessments of product proteins and impurities generated by Chinese hamster ovary (CHO) cells. Our presentation is necessarily brief, and although we address implications for analytics and specific assays, we focus primarily on application of proteomic methods to biomanufacturing processes.
Proteomics of CHO HCPs
LC-MS technology has matured rapidly over the past decade and has displaced two-dimensional gel electrophoresis to assume a dominant role in proteomics. Today, analysts often can identify and quantify hundreds to thousands of discrete protein species in a single sample, a capability that should serve outstandingly well in HCP analysis (4, 5). For some HCPs, methods even have been developed to detect levels as low as
0.1 ppm (6). However, access to seemingly comprehensive data sets can be highly misleading without adequate perspective on uncertainties that are inherent to proteomic analysis.
Converting the exquisite sensitivity of modern mass spectrometers into large data sets involves multiple steps both upstream and downstream of an LC-MS instrument (7). A sample must be exchanged into an appropriate buffer environment and digested by a protease before the resulting peptides are separated by one or two LC steps, with the eluting peaks then analyzed by MS. Some analyte components might be lost during sample preparation, whereas others could be obscured by coeluting LC peaks. And after LC-MS data have been acquired, they are interpreted using software packages in which selected parameter values can influence final outcomes.
Although all assays have limitations, LC-MS analysis of HCPs is especially susceptible to “false negatives,” in which HCP species are present in a sample but remain undetected. Amid substantial quantities of a product protein, peaks from MAb-derived peptides can obscure those from HCP-derived peptides such that the number of HCP species detected is reduced by more than an order of magnitude (8). To overcome such issues, researchers have developed new workflows that leverage depletion of MAbs in a sample to yield one or more low-MAb fractions in which individual HCPs can be detected more easily (6, 9, 10). However, complete elimination of false negatives is difficult to envisage.
HCP Clearance in Downstream Processing
That a MAb product is secreted into the cell-culture fluid is one of the strengths of CHO and related mammalian cell lines. However, many HCPs that must be removed during downstream processing are likewise secreted (11). Moreover, cell death and lysis during culture can release intracellular HCPs and more complex biophysical structures such as organelles. In principle, the most direct way of dealing with HCPs would be at their source, and proteomic analysis is indeed performed within the cell-culture context (12, 13). However, our emphasis below is on HCP clearance during purification of MAbs from harvested cell-culture fluid (HCCF) under typical cell-culture conditions (e.g., with titers of ~10 mg/mL).
A downstream process can achieve robust clearance of all impurity classes through multiple unit operations performed in series with orthogonal patterns of separation. For MAb biomanufacturing, the well-established platform process involves a capture step using protein A affinity chromatography, which is highly specific and typically achieves HCP log reduction values (LRVs) of ~3 log10 (2, 3). Remaining impurities, usually already at low concentrations, are then reduced to acceptable levels by additional polishing steps. Selection of unit operations (mainly of different chromatography modes) often is guided by heuristics regarding their effectiveness at removing different impurities (3). For instance, flow-through anion-exchange (AEX) chromatography is applied widely to reduce HCPs and hcDNA, but it is not considered to be effective at removing MAb aggregates, for which cation-exchange (CEX) chromatography and hydrophobic-interaction chromatography (HIC) are used more frequently.
Platform processes structured in such ways can reliably reduce overall HCP concentrations to low levels. This fact might seem inconsistent with the observation that damaging levels of individual HCPs sometimes persist in drug substances and drug products. One factor to bear in mind is the extremely low concentrations at which some enzymes can have deleterious effects on therapeutic proteins. A related consideration is the fairly long shelf-life expected for most biopharmaceuticals.
Often, multiple individual HCPs are detected in drug substances. In the absence of harmful effects on a product or patient, such persistence generally is considered to be benign. However, recurring issues arising from high-risk HCPs have prompted the biopharmaceutical community to investigate mechanisms that might contribute to persistence of individual HCPs through, for instance, a chromatography step. Three mechanisms have received the most attention.
Coelution: The most straightforward explanation for poor separation is that a given product protein and HCP behave nearly identically in the separation train, specifically in having similar binding and elution properties during chromatography (Figure 1). Although coelution is plausible within a single unit operation (14), its likelihood is much lower when considering all of the multiple, nominally orthogonal operations in a usual downstream train.
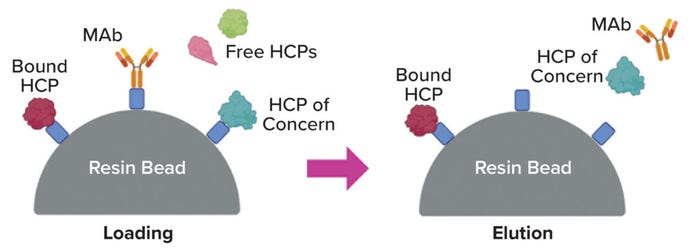
Figure 1: Coelution is a possible mechanism of host-cell protein (HCP) persistence through chromatography of a monoclonal-antibody (MAb) product (e.g., a protein A step), shown below from loading through elution (image created using BioRender software).
Product Association: A potential driver of persistence is noncovalent association between an HCP and product, with the complex having similar separation properties to those of the product (15) (Figure 2). Researchers have investigated the possibility of such “hitch-hiking” in various ways — e.g., by placing material from a null HCCF process (no MAb present) in direct contact with a MAb immobilized onto chromatographic particles (16, 17). Such studies have helped to identify dozens of HCPs that appear to bind with multiple MAbs — sometimes with most or all of those tested. Such observations could help explain not only the repeated finding of certain persisting HCPs, but also less predictable cases in which a certain high-risk HCP is found to be problematic for a particular MAb process.
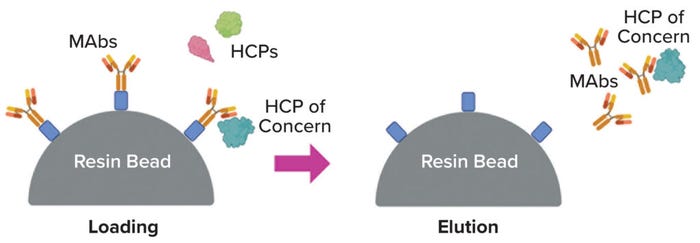
Figure 2: Product association as a potential mechanism for host-cell protein (HCP) persistence (image created with BioRender software).
Implicit in much of the product-association discussion is the notion that such binding is strong and specific — it is the high-affinity binding that reflects a (fortuitously) high degree of molecular complementarity between the proteins concerned. However, in a few reported cases, direct measurement of the MAb dissociation constants (KD values) indicated that the affinities are not particularly high (KD ~ 1 µM) (18, 19). In addition, most of the HCPs that appear to exhibit product association are among the most abundant ones in HCCF (19). Thus, the association seems to be driven less by high affinity than by mass action coupled with moderate affinity, possibly in multiple binding configurations (hence more accurately characterized as avidity).
Such observations suggest a situation in which product association indeed contributes to persistence of an HCP, with repeated reequilibrations leading gradually to its depletion. Reequilibration could occur, for instance, during a wash step in a chromatography process, and the conditions of the wash (e.g., pH or the presence of an excipient) might further modulate the dissociation constant of the complex.
Aggregates: Often called HMW species, aggregates long have been treated as a major class of product-related impurities to be cleared during downstream MAb processing (20) (Figure 3). Typically, they are regarded as MAb oligomers, and their properties during chromatography tend to be similar to those of a MAb monomer, including their capacity for persistence through protein A chromatography steps. About a decade ago, Gagnon et al. reported evidence that some aggregates can serve as carriers of HCPs and thereby might be responsible for HCP persistence in many cases (21). The writers hypothesized that such aggregates form around chromatin particles, in which histones are highly positively charged and DNA is highly negatively charged; other species, including HCPs and MAbs, would then bind promiscuously because of strong electrostatic attraction. Several subsequent studies have shown that pretreatment to remove aggregates can improve HCP clearance appreciably during protein A chromatography (22–24).
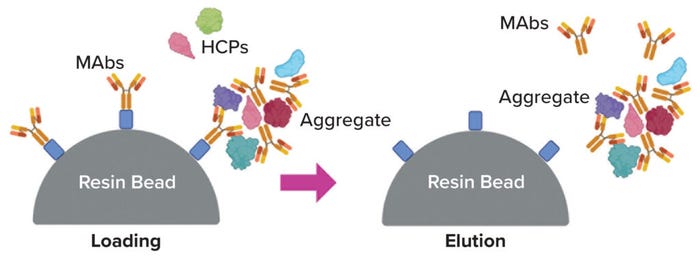
Figure 3: Aggregation as a potential mechanism for host-cell protein (HCP) persistence (image created with BioRender software).
More recent evidence supports the likely role of aggregates in HCP persistence. Proteomic analysis shows that MAb aggregates, classified somewhat arbitrarily into larger and smaller fractions (with radii up to ~50 nm and ~10 nm, respectively), contain many hundreds of different HCPs (8). Those include cellular-defense proteins such as chaperones, suggesting a possible origin for aggregates other than histones (25). Size-exclusion chromatography (SEC) analysis of protein-A fractions reveals that although free HCPs almost entirely flow through during column loading, aggregates bind similarly to — and evidently competitively with — MAb monomers, and a significant proportion of those aggregates coelute with the monomers (26). That finding supports the conventional wisdom that aggregates contain MAb molecules, but the proteomic data show that both large and small aggregates contain HCPs as well.
SEC fractionation of material from flow-through AEX presents a complementary picture (26). For AEX processes, conditions are chosen such that MAb monomers do not bind to resin beads, but results from SEC analysis show that such conditions limit binding of small aggregates, too, explaining why flow-through AEX is disfavored for aggregate clearance. In contrast, large aggregates may bind and be removed, making flow-through AEX effective for HCP clearance as a polishing step. The small aggregates appear to be more MAb-rich than the large ones are, so small aggregates more closely resemble the conventional view of HMW species as MAb oligomers (8).
Making Sense of HCP Persistence
The picture that emerges from such findings is helping researchers to explain the mechanisms underlying the fate of HCPs during downstream processing. Some moderately abundant HCPs appear to persist by product association, but they also seem to be depleted during operations such as wash steps that enable dissociation of MAb–HCP complexes and removal of freed HCPs. This model of HCP persistence is consistent with the observation that product association strength differs across product species, resulting in differing amounts of residual HCPs.
Aggregates, regardless of their origins, provide another likely and perhaps more widespread mechanism for HCP persistence. Available data clearly demonstrate that aggregates can elude clearance during protein A chromatography but that polishing steps such as flow-through AEX can remove them effectively. That aggregates are probably the principal vectors of HCP persistence is supported by a broad correlation between HCP numbers/concentrations and HMW content in tested material (26) and by correlation of HCP persistence with MAb aggregation propensity (27).
Such a model of HCP persistence muddies the customary distinction between product- and process-related impurities, the two categories into which HMW and HCP species are normally classified, respectively. Aggregates that contain HCPs represent both product- and process-related impurities; consequently, they can confound heuristics regarding the suitability of different unit operations for removing given types of impurities. For example, flow-through AEX typically is considered to be effective at removing HCPs but not HMW species. That notion seems to be inconsistent with the observation that large, HCP-containing aggregates are indeed cleared effectively. Resolution of that apparent conflict lies in the poor clearance of small aggregates, which might be biased heavily toward MAb oligomers even if they also include simple MAb–HCP complexes.
What do such findings tell us about the structure and operation of downstream processes to remove HCPs and other impurities? For MAbs, the established platform process generally is highly effective, but enhanced analytical support can help to identify areas of potential concern and improved control. Recent study results confirm what Gagnon et al. proposed a decade ago: that despite its effectiveness and dominance over other purification strategies, the Achilles heel of protein A chromatography is poor clearance of HCP-containing aggregates. Resins and complementary technologies that are modified to address such impurities could be beneficial. For modalities other than MAbs, questions remain about whether a given product and formulation are susceptible to damage from individual HCPs. Lessons learned from experiences with MAbs will provide both guidance and adaptable purification methods.
References
1 Jones M, et al. “High-Risk” Host Cell Proteins (HCPs): A Multi-Company Collaborative View. Biotechnol. Bioeng. 118(8) 2021: 2870–2885; https://doi.org/10.1002/bit.27808.
2 Shukla A, et al. Downstream Processing of Monoclonal Antibodies — Application of Platform Approaches. J. Chromatogr. B 848(1) 2007: 28–39; https://doi.org/10.1016/j.jchromb.2006.09.026.
3 Shukla A, et al. Evolving Trends in MAb Production Processes. Bioeng. Translat. Med. 2(1) 2017: 58–69; https://doi.org/10.1002/btm2.10061.
4 Falkenberg H, et al. Mass Spectrometric Evaluation of Upstream and Downstream Process Influences on Host Cell Protein Patterns in Biopharmaceutical Products. Biotechnol. Prog. 35(3) 2019: e2788; https://doi.org/10.1002/btpr.2788.
5 Oh Y, et al. Identification and Characterization of CHO Host-Cell Proteins in Monoclonal Antibody Bioprocessing. Biotechnol. Bioeng., submitted, 2023.
6 Yang F, et al. Versatile LC–MS-Based Workflow with Robust 0.1 ppm Sensitivity for Identifying Residual HCPs in Biotherapeutic Products. Anal. Chem. 94(2) 2022: 723–731; https://doi.org/10.1021/acs.analchem.1c03095.
7 Guo J, et al. Technical Advancement and Practical Considerations of LC-MS/MS-Based Methods for Host Cell Protein Identification and Quantitation To Support Process Development. mAbs 15(1) 2023: 2213365; https://doi.org/10.1080/19420862.2023.2213365.
8 Herman CE, et al. Analytical Characterization of Host-Cell-Protein-Rich Aggregates in Monoclonal Antibody Solutions. Biotechnol. Prog. 39(4) 2023: e3343; https://doi.org/10.1002/btpr.3343.
9 Huang L, et al. A Novel Sample Preparation for Shotgun Proteomics Characterization of HCPs in Antibodies. Anal. Chem. 89(10) 2017: 5436–5444; https://doi.org/10.1021/acs.analchem.7b00304.
10 Nie S, et al. Simple and Sensitive Method for Deep Profiling of Host Cell Proteins in Therapeutic Antibodies By Combining Ultra-Low Trypsin Concentration Digestion, Long Chromatographic Gradients, and Boxcar Mass Spectrometry Acquisition. Anal. Chem. 93(10) 2021: 4383–4390; https://doi.org/10.1021/acs.analchem.0c03931.
11 Kumar A, et al. Elucidation of the CHO Super-Ome (CHO-SO) By Proteoinformatics. J. Proteome Res. 14(11) 2015: 4687–4703; https://doi.org/10.1021/acs.jproteome.5b00588.
12 Park JH, et al. Proteomic Analysis of Host Cell Protein Dynamics in the Culture Supernatants of Antibody-Producing CHO Cells. Sci. Rep. 7, 2017: 44246; https://doi.org/10.1038/srep44246.
13 Hamaker NK, Min L, Lee KH. Comprehensive Assessment of Host Cell Protein Expression After Extended Culture and Bioreactor Production of CHO Cell Lines. Biotechnol. Bioeng. 119(8) 2022: 2221–2238; https://doi.org/10.1002/bit.28128.
14 Levy NE, et al. Host Cell Protein Impurities in Chromatographic Polishing Steps for Monoclonal Antibody Purification. Biotechnol. Bioeng. 113(6) 2016: 1260–1272; https://doi.org/10.1002/bit.25882.
15 Shukla AA, Hinckley P. Host Cell Protein Clearance During Protein A Chromatography: Development of an Improved Column Wash Step. Biotechnol. Prog. 24(5) 2008: 1115–1121; https://doi.org/10.1002/btpr.50.
16 Aboulaich N, et al. A Novel Approach To Monitor Clearance of Host Cell Proteins Associated with Monoclonal Antibodies. Biotechnol. Prog. 30(5) 2014: 1114–1124; https://doi.org/10.1002/btpr.1948.
17 Levy NE, et al. Identification and Characterization of Host Cell Protein Product-Associated Impurities in Monoclonal Antibody Bioprocessing. Biotechnol. Bioeng. 111(5) 2014: 904–912; https://doi.org/10.1002/bit.25158.
18 Ranjan S, et al. Investigation of Cathepsin D–MAb Interactions Using a Combined Experimental and Computational Tool Set. Biotechnol. Bioeng. 116(7) 2019: 1684–1697; https://doi.org/10.1002/bit.26968.
19 Oh Y, et al. Factors Affecting Product Association as a Mechanism of Host-Cell Protein Persistence in Bioprocessing. In preparation, 2023.
20 Vázquez-Rey M, Lang DA. Aggregates in Monoclonal Antibody Manufacturing Processes. Biotechnol. Bioeng. 108(7) 2011: 1494–1508; https://doi.org/10.1002/bit.23155.
21 Gagnon P, et al. Nonspecific Interactions of Chromatin with Immunoglobulin G and Protein A, and Their Impact on Purification Performance. J. Chromatogr. A 1340, 2014: 68–78; https://doi.org/10.1016/j.chroma.2014.03.010.
22 Nian R, et al. Advance Chromatin Extraction Improves Capture Performance of Protein A Affinity Chromatography. J. Chromatogr. A 1431, 2016: 1–7; https://doi.org/10.1016/j.chroma.2015.12.044.
23 Ichihara T, Ito T, Gillespie C. Polishing Approach with Fully Connected Flow-Through Purification for Therapeutic Monoclonal Antibody. Eng. Life Sci. 19(1) 2019: 31–36; https://doi.org/10.1002/elsc.201800123.
24 van de Velde J, et al. Chromatographic Clarification Overcomes Chromatin-Mediated Hitch-Hiking Interactions on Protein A Capture Column. Biotechnol. Bioeng. 117(11) 2020: 3413–3421; https://doi.org/10.1002/bit.27513.
25 Oh YH, et al. Characterization and Implications Of Host-Cell Protein Aggregates in Biopharmaceutical Processing. Biotechnol. Bioeng. 120(4) 2023: 1068–1080; https://doi.org/10.1002/bit.28325.
26 Herman CE, et al. Behavior of Host-Cell-Protein-Rich Aggregates in Antibody Capture and Polishing Chromatography. J. Chromatogr. A 1702, 2023: 464081; https://doi.org/10.1016/j.chroma.2023.464081.
27 Luo H, et al. Formation of Transient Highly-Charged MAb Clusters Strengthens Interactions with Host Cell Proteins and Results in Poor Clearance of Host Cell Proteins By Protein A Chromatography. J. Chromatogr. A 1679, 2022: 463385; https://doi.org/10.1016/j.chroma.2023.464081.
Corresponding author Abraham M. Lenhoff is Allan P. Colburn Professor in the Department of Chemical and Biomolecular Engineering at the University of Delaware, 150 Academy Street, Colburn Laboratory, Newark, DE 19716; [email protected]. Chase E. Herman is an investigator in biopharmaceutical drug substance development at GSK.
You May Also Like





