Replacing Reverse-Phase Chromatography for Mass Spectrometry: Is Salt-Free Size-Exclusion Chromatography Ready?

A Waters Acquity column and an Agilent Q-ToF system were used in this study.
Protein mass is often determined using ultraperformance liquid chromatography (UPLC) coupled with electrospray-ionization mass spectrometry (UPLC/ ESI MS or simply LC-MS). A UPLC system equipped with an ultraviolet (UV) detector serves as an assisting vehicle to deliver purified and separated protein molecules to the mass analyzer. Reserved-phase chromatography (RPC) is the most common chemistry chosen to serve this purpose. For sample purification, not only does RP-UPLC use salt-free mobile phases that are amenable to MS, but it also can efficiently remove buffers, salts, and other sample components that otherwise would interfere with a mass analysis. In addition to sample purification, RP-UPLC has sufficient separation capability to readily separate heavy-chain (HC) and light-chain (LC) antibody domains under reducing conditions (1).
However, RP-UPLC/MS has limitations in protein mass analysis. The technology separates proteins based on their hydrophobicity, but molecules with different sizes that consist of the same amino-acid sequence (e.g., protein dimers or half antibody fragments) may show similar hydrophobicity. Also, as we discuss below, RP-UPLC cannot resolve the HC and LC peaks of a reduced antibody based on retention time. That can potentially cause problems related to deconvolution of mass spectra.
Unlike RPC, size-exclusion chromatography (SEC) separates molecules simply based on their size differences (2). It is the predominant method used by the biopharmaceutical industry to detect antibody aggregation and degradation. SEC can remove sample matrix components and separate proteins. Coupling SEC with MS could be a favorable alternative for overcoming the limitations of RP-UPLC/MS.
In current biopharmaceutical industry practice, however, SEC lags behind RPC in MS applications for a number of reasons. Traditional SEC uses a high-salt mobile phase that is incompatible with ESI-MS. Even when converted to an acetonitrile– water solution as a salt-free mobile phase, traditional SEC columns are incompatible with UPLC. To use such a column for MS analysis, a post- column flow splitter has been recommended for implementation before MS analysis (3). But that can reduce assay sensitivity.
The recent introduction of a <2-µm UPLC SEC column that is compatible with salt-free mobile phases makes it possible for SE-UPLC/MS to become a viable and simple method for determining protein mass. In our study, we evaluated this technique for antibody mass determination under reduced and intact conditions, then
compared results with those obtained using RP-UPLC/MS.
Materials and Methods
We evaluated SE-UPLC/MS using a 1.7-mm ID, 4.6 × 300-mm Acquity UPLC BEH200 SEC column from Waters Corporation (part #186005226) installed in a 6530 quadrupole time-of-flight (Q-TOF) LC/MS system from Agilent Technologies. Unless otherwise
specified, SE-UPLC involved an isocratic elution at 0.3 mL/minute with a salt-free mobile phase consisting of 30% (v/v) acetonitrile/0.05% (v/v) trifluoroacetic acid (TFA) in an aqueous solution. We set the UPLC-SEC column temperature at room temperature. For comparison, RP-UPLC/MS was performed on the same Agilent Q-TOF LC/MS system using a 1.7-mm ID, 2.1 × 100-mm Zorbax RPHD 300SB-C8 column from Agilent Technologies (part #858750- 906). We set the C8 column at 75 °C. The RP-UPLC process involved a gradient of 0.1% (v/v) formic acid in aqueous solution as mobile phase A and 0.1% (v/v) formic acid in acetonitrile as mobile phase B at 0.5 mL/minute. For 0–3 minutes, we applied 5–20% B; from 3 to 13 minutes, 20–80% B; from 13 to 15 minutes, 80% B; and from 15 to 20 minutes, 5% B.
For both UPLC methods, we injected 10 µg of protein sample and monitored it by UV at 214 nm for the reduced and 280 nm for the intact samples. The Agilent 3650 QTOF
system settings were as follows: positive ion polarity; 325 °C drying gas at 10 L/ minute; 60-psi nebulizer; 5,500-V Vcap, 160-V fragmentor, and 65-V skimmer; with four MS scans/second at 300–3,200 m/z. Reference correction was set for one point at 922 m/z, and the acquisition mode was extended dynamic at 2 GHz. We performed the MS analysis using Agilent Masshunter software (version B.05), which included Agilent Bioconfirm software for data processing.
We chose two humanized monoclonal antibodies (MAb1 and MAb2) for this study. For reduced MAb1 and MAb2, we incubated samples with 6.3 M guanidine and 0.024 M dithiothreitol at 37 °C for 30 minutes. We also degraded a MAb1 sample with thermal stress at 55 °C for 48 hours.
Results and Discussion
We analyzed intact MAb1 (an aglycosylated antibody) by SE-UPLC/MS in parallel with RP-UPLC/MS as described above. The upper panels of Figure 1 show total-ion (TI) SEC and TI RPC results for intact MAb1, and the lower panels show corresponding UV (280-nm) chromatograms.
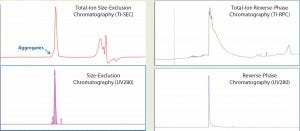
Figure 1: Total-ion (TI) size-exclusion chromatography (SEC, left) and reverse-phase chromatography (RPC, right) of MAb1 are listed in the upper panels, and their corresponding UV (280 nm) chromatograms are given in the lower panels. Both ultraperformance liquid chromatographic methods provide a single peak for mass analysis. But unlike RPC, the retention segment of an SEC peak would provide additional aggregate information.
Both analytical methods could resolve a single, salt-free peak for protein mass determination. However, a closer look reveals that the SEC peak has a peak shoulder on the left side of the main peak, whereas the RPC does not.
Because of the different separation mechanisms, that left peak shoulder could contain a molecular species of different size from that in the main peak. To verify whether the SE-UPLC/MS method could distinguish the mass difference between those peak segments, we divided the MAb1 SEC peak (Figure 1) into a left shoulder and a main peak to analyze them separately for their masses. Figure 2 shows the mass results. As the upper-panel picture inserts show, ESI mass spectrums (ESI MS) derived from the shoulder differed significantly from the main peak, implying that the shoulder species was not the same molecule as that in the main peak.
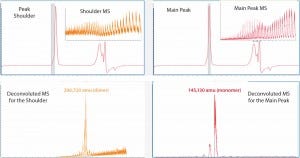
Figure 2: The Mab1 SEC peak in Figure 1 was analyzed for mass identification. The peak shoulder (left) and main peak (right) were processed separately. Inserts show ESI mass spectra and corresponding deconvoluted mass spectra are shown in the lower panels.
Next we deconvoluted the two ESI MS datasets from the left shoulder and main peak separately using Agilent Bioconfirm software and calculated their mass values. Deconvoluted MS indicated that the estimated mass of the left-peak shoulder species was 290,720 amu, which was two times larger than the mass value of 145,130 amu derived from the main peak. So the left shoulder represented the antibody dimer. Hence, the SE-UPLC/MS method could be used to confirm the presence of antibody dimer in a sample.
By contrast, the RP-UPLC/MS could not separate any dimer fraction (Figure 1). Those results estimated the mass of the main peak for the intact MAb1 to be 145,130 amu (deconvoluted MS not shown), which was identical to the SE-UPLC/MS result.
Thermal Degradation: In a separate experiment to determine whether the SE-UPLC/MS could identify a protein mass change due to a particular process, we subjected an intact MAb1 sample to thermal treatment and analyzed it using SE-UPLC/MS (Figure 3).
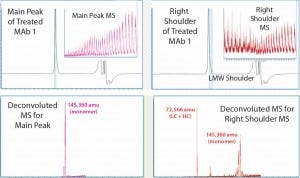
Figure 3: MS results for MAb1 treated at 55 C — with the main peak (left) and right peak shoulder (right) processed separately; inserts show ESI mass spectra, and lower panels show corresponding deconvoluted mass spectra.
Comparing the results with those in Figure 2 for untreated MAb1, we found that in the SEC of thermally treated MAb1, the left-peak shoulder of dimer dwindled significantly and a right-peak shoulder emerged. To process mass-shift data from the thermal treatment, we divided the right shoulder of the MAb1 SEC peak from the main peak and analyzed it separately to find its mass. Again, the ESI MS derived from the shoulder gave a different profile from what was derived from the main peak.
Deconvoluted MS indicated that the estimated mass for the main peak was 145,360 amu, whereas the right shoulder contained mainly two species that were not separated from one another. We calculated the mass of the main species to be 72,566 amu, which appeared to be half of a monomer (a combination of one LC with one HC). And we estimated the mass of the minor species to be 145,360 amu, which was the same as that estimated for the main peak, so it should also be a monomer. Hence, SE-UPLC/MS also separates half-antibody fragments for identification. The monomer size of the thermally treated MAb1 was slightly larger than that of the untreated antibody, which might be attributed to potential protein oxidation during thermal treatment.
Reducing Conditions: To analyze both intact and reduced antibody samples, we used a salt-free mobile phase for SEC-UPLC. A mobile phase consisting of 30% (v/v) acetonitrile and 0.05% (v/v) trifluoroacetic acid (TFA) in aqueous solution was compatible with the water–acetonitrile gradient with 0.1% (v/v) formic acid we used with the RP-UPLC for comparison in this study. Results showed that the presence of 0.05–0.1% (v/v) TFA in the mobile phase was a critical factor to maintaining and improving SEC column performance under salt-free conditions (4). TFA is known as an ion-pairing reagent for protein. Many relevant studies have used formic acid as a mobile-phase additive in SEC, but we found adding 0.05% (v/v) formic acid into the salt-free mobile phase had no effect on column performance.
For reduced antibody samples, we chose MAb1 and MAb2 for this study. The distinct difference between the two MAbs was that unlike aglycosylated MAb1, MAb2 had normal IgG glycosylation. We used treatment with 6.3 M guanidine and 0.024 M dithiothreitol at 37 °C incubation to reduce the two MAbs to their corresponding HC and LC components. Figure 4 shows SE-UPLC and RP-UPLC results for reduced MAb1 and MAb2. As the figure shows, both UPLC methods could separate HC and LC for mass analysis. However, SE-UPLC had the better resolution — and in particular, better consistency than RP-UPLC.
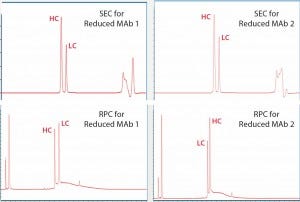
Figure 4: Upper panels show UV280 SEC of reduced Mab1 (left) and reduced MAb2 (right), and lower panels show UV280 RPC for the same samples. Heavy- chain (HC) and light-chain (LC) peaks are identified and annotated.
Data Processing: Size differences between the HC and LC domains makes SEC peak elution patterns predictable, which is favorable, by narrowing the predicted mass range to prevent redundancy of deconvolution for data processing. On the other hand, the RPC peak elution pattern is unpredictable because it depends on hydrophobicity differences between the HC and LC domains. The HC peak could be eluted either before or after the LC peak, as we observed respectively in our MAb1 and MAb2 analyses, which can cause potential deconvolution problems.
Another advantage of the SE-UPLC/MS method for reduced antibody samples is that it can identify potential size modifications of the HC or LC domains (5): e.g., identifying HC–HC clips with SE-UPLC/MS. Estimated mass values of the HC and LC domains from our SE-UPLC/MS results were basically identical to those estimated based on RP-UPLC/MS. For example, the estimated mass values according to SE-UPLC/MS for MAb2 were 51,878 amu for the HC and 24,001 amu for the LC — compared with 51,880 amu and 24,001 amu obtained, respectively, using the RP-UPLC/MS.
Table 1: SE-UPLC consistency results for MAb1
(a Coefficient of variation b Coefficient of determination)
CVa (n = 3) | ||||
MAb1 | Peak Area | Retention | Loading Range | RSQb |
Heavy Chain | 0.7% | 0.5% | 4−20 μg/injection | 0.98 |
Light Chain | 0.8% | 0.5% | 4−20 μg/injection | 0.99 |
Monomer | 2.9% | 2.2% | 4−20 μg/injection | 1.00 |
We also tested the separation consistency of an MS-friendly SE-HPLC method. Table 1 lists precision and linearity results obtained for MAb1 with 4–20 µg per injection. This method showed excellent repeatability for retention time and peak area as well as linearity of response. And we concluded that SE-UPLC/MS may well offer a superior alternative method for LC-MS analysis of proteins.
References
1 Rehder DS, et al. Reverse-phase Liquid Chromatography/Mass Spectrometry Analysis of Reduced Monoclonal Antibodies in Pharmaceutics. J. Chromatogr. A 1102 (1–2) 2006: 164–175.
2 Barth HG, Boyes BE, Jackson C. Size-Exclusion Chromatography. Analyt. Chem. 66(12) 1994: 595R–602R.
3 Gadgil H, et al. SEC–MS Analysis of Aggregates in Protein Mixtures. The Applications Book. Waters Corporation: Milford, MA, September 2003.
4 Liu H, Gaza-Bulseco G, Chumsae C. Analysis of Reduced Monoclonal Antibodies Using Size Exclusion Chromatography Coupled with Mass Spectrometry. J. Am. Soc. Mass Spectrom. 20, 2009: 2258–2264.
5 Chakraborty A, Chen W, Mazzeo J. A Generic On-Line UPLC-SEC/UV/MS Method for the Analysis of Reduced Monoclonal Antibodies. App. Solutions Biopharmaceut. September 2012: 61–62
Corresponding author Xi Lin and Jichao Kang are in the analytical and formulation department of Gallus Biopharmaceuticals, LLC, 201 College Road East, Princeton, NJ 08540; 1-609- 919-3300; [email protected].
You May Also Like





