Voices of Biotech
Podcast: MilliporeSigma says education vital to creating unbreakable chain for sustainability
MilliporeSigma discusses the importance of people, education, and the benefits of embracing discomfort to bolster sustainability efforts.
July 1, 2008

Figure 1.
Established in 1997, integrated Genomics (IG) provides a full range of products and services to support research in microbial genomics, metabolic analysis, and engineering. Our flagship genome analysis system, ERGO™, provides a bioinformatics toolkit enabling better and faster identification of gene function and pathways across organisms, which is evident within more than 100 scientific publications in top peer-reviewed journals (www.integratedgenomics.com/igpubs.html).
The ERGO in silico metabolic discovery and analysis tool set is the world’s largest commercially available database of microbial genomes coupled with a state-of-the-art analytical tool set. More than 900 bacterial genomes have been curated manually and associated with a network of biochemical pathways. Currently, ERGO contains more than 3.6 million open reading frames, of which 2.5 million have functional assignments and 6,500 metabolic pathways grouped into subsystems, all linked together with data mining and analysis tools. ERGO also supports integration and examination of gene expression data in the framework of both primary genomic data and the pathway network. The metabolic architecture of each microorganism can be readily analyzed in its physiological context.
Leaders in Microbial Metabolic Engineering
Whole-genome sequencing, bioinformatics, and metabolic reconstruction have not only accelerated product optimization and improved microbial strain growth, but also have facilitated the creation of new strains, thus increasing the pace of commercialization. IG provides technical expertise across all stages of the ME process including examination of gene expression, strain design, strain construction, screening selection, and growth optimization/fermentation/production (Figure 1).
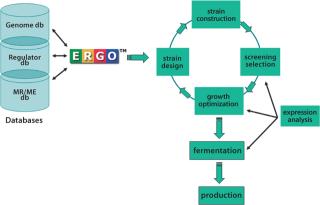
Examination of gene expression and key chemical compound production in the context of a metabolic pathway network is the first step in product engineering. For example, scientists at IG have examined the expression of all the metabolic genes of Escherichia coli MG1655 grown in LB-broth under normal laboratory conditions and compared the resulting data with compound metabolism studies to determine the relationship between gene expression and carbon use over time (1, 2). This established a set of gene targets that can be manipulated during strain design (Figure 2).

Strain design involves the alteration of existing pathway networks to improve product yield. Using metabolic network blueprints, gene and pathway information can be leveraged to engineer a production strain by standard molecular “cut-and-paste” techniques. The resulting biochemical maps help quickly identify and evaluate novel genes as well as highlight metabolic bottlenecks, pathway branch-points, and redundancies so carbon flow can be appropriately channeled. Pathway engineering can then be implemented by “knocking-out” genes in pathways that drain the flow of substrate to an undesired product or by “knocking-in” genes necessary to redirect substrate flow toward the desired product.
The final step in ME (growth optimization/fermentation) involves more “downstream” tasks, from development of suitable microbial growth media based on a detailed nutritional profile of the engineered organism (3) to laboratory-scale fermentation suitable for pilot scale-up (1, 2).
Integrated Genomics is a world leader in metabolic engineering, providing one-stop, full-service genomics solutions. Focused on microbial genomics, ERGO™ integrates proprietary functional genomic data, metabolic reconstructions, expression profiling, and biochemical and microbiological data with publicly available information to provide better and faster identification of gene function across all organisms.
1.) Baev, MV. 2006a. Growth of Escherichia coli MG1655 on LB Medium: determining Metabolic Strategy with transcriptional Microarrays. Appl. Microbial. Biotechnol. 3:323-328.
2.) Baev, MV. 2006b. Growth of Escherichia coli MG1655 on LB Medium: Monitoring Utilization of Sugars, alcohols, and organic acids with transcriptional Microarrays. Appl. Microbial. Biotechnol. 3:310-316.
3.) Bhattacharyya, A. 2002. Draft Sequencing and Comparative Genomics of Xylella fastidiosa Strains reveal Novel Biological insights. Genome Res. 10:1556-1563.
You May Also Like