Use of CRISPR and Other Gene-Editing Tools in Cell Line Development and Engineering
September 23, 2020
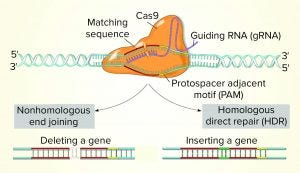
Figure 1: CRISPR-Cas9 (www.stockadobe.com)
While the role of biologics in treating human diseases has evolved dramatically over the past decade, so has genetic engineering. Rational genetic engineering to enhance biotherapeutic proteins has become a reality catalyzed by publication of the genome sequences of multiple Chinese hamster ovary (CHO) cell lines. Novel “designer” CHO cells modulate posttranslational modifications (PTMs) of recombinant proteins by genome editing, and it is now possible to knock-in or knock-out genes of yeast and mammalian cells precisely (within one DNA base pair), quickly, and efficiently. Eventually, such techniques will be used in the development of cost-effective recombinant therapeutic proteins.
The gene-editing tool based on bacterial clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated protein 9 (Cas9) has improved genome editing dramatically, making it faster, easier, less expensive, and more efficient than before. This system needs only one protein and one RNA molecule to achieve RNA-programed DNA cleavage. That capability has enabled CHO researchers to elucidate the mechanistic basis behind the high-level protein production and product quality attributes (PQAs) of interest. Currently, the emphasis in CHO gene editing is directed toward expanding product diversity as well as controlling and improving product quality and yields. Although routine PTM optimization across a cell’s glycosylation “machinery” is not yet a reality, researchers hope to eventually be able to combine the expression from nonmammalian hosts with human-like antibody glycosylation performed in engineered yeast or even plants.
But the use of promoter engineering to achieve precision transcriptional control for CHO-cell synthetic biotechnology could be achieved in the near future. Similarly, multiplexed genome editing (simultaneous targeting of multiple related or unrelated targets) could enable the examination and manipulation of whole genomes or protein networks.Thus, it has revolutionized knockout and knockin events and has played a crucial role in the understanding of the genomes of mammalian cells and the use of yeasts for metabolic engineering. Such editing capabilities also have enabled high, stable, and consistent expression of transgenes encoding complex therapeutic proteins such as monoclonal antibodies (MAbs).
Posttranslational Modifications
Advanced gene-editing technologies such as CRISPR-Cas9, transcription activator-like effector (TALE) nucleases, and RNA interference (RNAi) tools are complemented by the combination of next-generation sequencing with systems biotechnology to facilitate further enhancements in cell glycosylation processing. Such tools will enable cell engineers to make highly refined and targeted modifications to fast-track the processing capability of cells. That will bring consistent improvements in yield and cell quality as well as increased process efficiency, thus leading to increased affordability for future healthcare needs.
Cell engineers are witnessing a proliferation of enabling tools as molecular and cell biologists continue to develop sophisticated techniques to decipher attributes that are critical to drug-product quality. Improving the glycosylation profiles of biologics enhances product bioactivity and quality, and N-linked glycosylation plays a crucial role in the efficacy of therapeutic proteins. Proven therapeutic efficacy of many glycoengineered proteins has stimulated the development of novel optimized expression systems (e.g., mammalian cells) for producing nonfucosylated antibodies.
It is now becoming feasible to produce material rapidly for pharmacology, formulation, and toxicology studies without having to establish a stable cell line. Such capability can shorten timelines for stable cell line development from over six months to about three weeks, thus enabling a generation of stable clones at the research stage. At the same time, different production systems for glyco-optimized proteins (e.g., yeast) already have been engineered to produce the main steps of the human N-glycosylation pathway and thus have enabled biobetter versions of therapeutic MAbs.
Application of CRISPR-Cas9 tools has led to multiplexed-knockout phenotypes. Similarly, upcoming achievements with (small) noncoding RNAs in CHO cells also could support functional genomics efforts to enhance producer cells. That would expand the list of engineering targets significantly (e.g., as achieved with obinutuzumab, a humanized and glycoengineered anti-CD20 MAb, with bisected afucosylated Fc-region carbohydrates and GlycoMAb technology, from GlycArt-Roche).
Not long ago, CHO cells were considered “black boxes” because researchers lacked the genomic information of those cells. That hindered efforts to understand the molecular basis of high-quality production of recombinant proteins in CHO cells. Notably, impressive progress in CHO cell culture technology has been achieved by empirical approaches such as screening and process optimization, thus enabling high yields (10 g/L and more). However, such yields often are achieved only by certain types of cell lines, and a high degree of variability in recombinant CHO cells requires laborious and expensive processes to select the best clone for the production of each new therapeutic candidate.
Traditional recombinant cells are based on random plasmid integration of an expression cassette in the host genome. Transgenes are likely to be inserted in heterochromatin regions, which results in very weak gene-expression levels. So a substantial number of clones (generally multiple hundreds to a few thousand) must be screened to find those rare cells with a stably integrated plasmid in highly transcriptionally active chromatin regions (“hotspots”).
Sequencing efforts have resulted in availability of additional genomic sequence data for different CHO cell lines. Moreover, an active effort is underway in the CHO cell-line community to refine the genome assembly and annotation for the Chinese hamster. In the past 20 years, several molecular and cellular biology tools have been developed for targeted-site gene integration for eventually minimizing the randomness of gene insertion and increasing the predictability of high transgene expression.
Genome Editing
First-Generation Genome-Editing Tools: Several recombinases have been used for targeted site approaches, with Cre–lox and Flp–FRT being the most commonly used. Recombination-mediated cassette exchange (RMCE) technology also is attracting interest for targeted gene insertion. That technology has increased success rates and reduced timelines for the generation of stable industrial-grade CHO cell lines expressing MAbs reliably.
Second-Generation Genome-Editing Tools: This group includes endonucleases such as zinc finger nucleases (ZFN), meganucleases, transcription activator-like effectors nucleases (TALEN), and CRISPR-Cas9. Both ZFN and TALEN technologies rely on the ability to customize a DNA-binding domain for a specific sequence (the targeted sequence for cleavage) combined to a nuclease effector domain. CRISPR-Cas9 technology already has been validated for use with CHO cells and has been implemented to reduce production variability among clones. However, ZFN, TALEN and CRISPR-Cas9 technologies are used mostly for gene-specific knockout.
One critical challenge is to identify hotspots in a host genome that enable good expression levels and stability. Such specific integration might not be a one-size-fits-all approach because some therapeutic proteins could require a particular level of expression to fold correctly or to present adequate quality attributes (e.g., glycosylation and proteolytic processing). The discovery of additional naturally occurring RNA-guided nucleases offers increased targeting flexibility.
Some researchers also have engineered Cas9 enzymes to exhibit relaxed protospacer-adjacent motif (PAM) specificities. Such crucial advances expand the number of target loci that are amenable to RNA-guided genome editing. Other scientists have engineered Cas9 nuclease and managed to increase its DNA specificity dramatically. Thanks to the fusion of dead Cas9 (dCas9) to a cytidine deaminase enzyme that operates on ssDNA, “base editing” now can be used to generate point mutations in genomes.
Remarkably, CHO cells showed increased resistance to apoptosis after successful simultaneous (multiplex) disruption of fucosyltransferase 8 (FUT8), Bcl-2 associated X-protein (BAX), and Bcl-2 homologous antagonist killer (BAK) genes by CRISPR-Cas9 (1). The ZFN knockout approach achieved specific deletion of the glutamine synthetase (GS) and the dihydrofolate reductase (DHFR) genes in CHO cells, thus improving the selection stringency of the generated cell lines. Finally, both ZFN and a CRISPR-Cas9 approaches also were used for FUT8 gene-specific knockout. The resulting cell lines completely abolished fucosylation on the Fc domain of immunoglobulin G (IgG).
A less commonly used tool is mammalian artificial chromosome expression (ACE). This minigenome serves as an autonomous genetic element that replicates with cells. Its DNA sequence is customizable with different regulating elements that could help its expression.
The PiggyBac transposon system (System Biosciences) uses an efficient transposase purified from the cabbage looper moth (Trichoplusia ni) to integrate the gene of interest into a host genome. It has shown to improve yields for stable production of antibodies in CHO cell lines (2).
Genome-Editing Achievements: Introduction of additional N-glycan target sites into desired positions on the protein backbone by genetic mutation has been used to create glycoproteins with enhanced levels of glycosylation (overexpression of sialyltransferases and other glycosyltransferases, inhibition of sialidases) and sialylation. That has extended serum halflives and improved in vivo activity (3). (N-glycans also can be crucial for protein folding.) Thanks to a comprehensive ZFN knockout screen of glycosyltransferase genes and the identification of key genes that control decisive steps in N-glycosylation in CHO cells, it is now possible to provide homogeneous glycoforms.
Overexpression of B-cell lymphoma 2 (Bcl-2) or B-cell lymphoma-extra large (Bcl-xL) has shown precise and efficient inhibition of apoptosis in recombinant CHO cell cultures by enhancing the culture’s longevity, cell viability, and endurance under environmental stresses. Consequently, that renders greater yields of therapeutic protein, which is a definite economic advantage in the biopharmaceutical industry. Similarly, down-regulation of caspases (e.g., caspase-8 and -9) and knockouts of proapoptotic genes (e.g., BAX and BAK) enhance the viability of both batch and fed-batch cultures. (Down-regulation of such genes can be achieved with various genome-editing techniques such as ZFNs, TALENs, and Cas systems).
CRISPR Improvements to CAR T Cells, by Maribel Rios |
|---|
Chimeric antigen receptor (CAR) T-cell technology involves transducing targeted T cells with a T-cell receptor that enables selective targeting of tumor cells in a manner that they’re not naturally inclined to do. The use of chimeric antigen receptor (CAR) T cells for therapeutic purposes (e.g., anticancer) already is prevalent in disease research. Automation of bioprocesses using CAR T cells also has been explored. But until 2017, the connection between CAR T cells and CRISPR technology had not been established fully. One group that helped bridge those two technologies is the research team at the Memorial Sloan Kettering Institute Center for Cell Engineering. Directed by Michel Sadelain, the group has been an integral part of CAR-T therapy research, particularly targeting the CD19 molecule on white blood cells. CD19 is expressed on cells found in B-cell leukemia and resides on the cell surface, thereby making it available for antibody access. In 2017, Sadelain’s group published a study showing that directing a CD19-specific CAR to the T-cell receptor alpha constant (TRAC) locus increased T-cell potency. The modified cells “vastly outperformed conventionally generated CAR T-cells in mouse models of acute lymphoma leukemia,” with human trials planned (1). CAR T-cells typically are made using retroviruses, but they insert CAR genes into random loci in genomes. CRISPR can be used to deliver CAR genes to specified targets in genomes. And current research is exploring the use of CRISPR–Cas9 editing to create next-generation CAR T-cell products, including “universal” CAR T-cells, making these cells more potent and controllable (2). References1 Eyquem J, et al. Targeting a CAR to the TRAC Locus with CRISPR/Cas9 Enhances Tumour Rejection. Nature 543(7643) 2017: 113–117; https://doi.org/10.1038/nature21405. 2 Chenggong L, Mei H, Hu Y. Applications and Explorations of CRISPR/Cas9 in CAR T-Cell Therapy. Brief. Funct. Genom. 19(3) 2020: 175–182; https://doi.org/10.1093/bfgp/elz042. |
Therapeutic Protein Expression Regulation By Smart Promoters and Epigenomic Reprogramming
In the near future, research laboratories will be able to design transcriptional control systems and synthetic promoters, control the timing of gene expression at will by trigger-inducible transcription factors (TF), and protect against chromatin silencing. Researchers already have begun to edit the epigenome to alter regulation of a target gene. Moreover, effector fusions can be used to expand the repertoire of genome engineering modalities achievable using Cas9. And nonediting CRISPR tools are helping researchers to screen engineered cells for phenotypes of interest quickly and precisely.
CRISPR Intellectual Property Landscape and the Gene-Editing Innovation Roadmap
There is some risk that patent filings by academic institutions claiming critical components of the CRISPR-Cas9 technology could deter or slow down the development and use of the technology. However, research organizations and universities can establish a workable balance between access and control for essential research tools through sound management of intellectual property (IP). (For example, when the Cohen–Boyer patents were granted, Stanford University created a pioneering licensing program that provided a predictable legal framework for using its inventions. Nonexclusive licenses were made available to both companies and academic institutions. In fact, patent protection has played a significant role in the development of the technology to produce recombinant DNA in bacteria).
It has been widely publicized that several organizations have been filing patents over fundamental parts of the CRISPR-Cas9 system. Commercial assignees Dow AgroSciences and DuPont Nutrition Science together hold 33 inventions, all of which are related to crop and animal agriculture (genome-editing crop and weeds) and food (dairy industry) applications of the technology. In addition, the French company Cellectis holds rights on a broad patent for gene editing of cells in vitro. Academic institutions, through their licensing and spinoffs, are primarily in control of medical applications of CRISPR-Cas (e.g., The Broad Institute of Harvard University and the Massachusetts Institute of Technology (MIT) to Editas) with commercial partners (University of California at Berkeley to Caribou Biosciences and sublicensed to Intellia and Novartis).
It’s interesting that most patent holders appear to be pursuing a strategy of keeping an international option open for their portfolios. Fortunately, signs indicate that the pace of discovery and development of CRISPR is likely to continue, with a high probability of further improvements. The Broad Institute and MIT are building a portfolio on those principles and diversifying it.
Other gene-editing technologies might emerge to compete with or possibly even displace CRISPR-Cas (see “Alternative Gene-Editing Methods” box). Follow-on breakthroughs are likely to take center stage. Current patent holders will have difficulty pursuing restricted access to CRISPR-Cas9 for research use because it already is widely used in academic laboratories. The Broad Institute, MIT, and UC Berkeley already offer free use of the technologies they control for academic research purposes through Addgene, a nonprofit organization. Addgene uses the general terms of the Uniform Biological Material Transfer Agreement (UBMTA), which indicates that discoveries are owned by the recipient of the biological material and can be licensed for commercial use.
Inevitably, some of those new commercial applications could fall within the broadly drafted claims of the original patent and therefore require a second, commercial license. Inventors of follow-on applications using a CRISPR-Cas technology are likely to need a commercial sublicense from the respective exclusive commercial licensee that controls that technology (e.g., Editas, Caribou, Intellia, CRISPR Therapeutics, and Cellectis/Calyxt) rather than from the originating university.
Alternative Gene-Editing Technologies, by Maribel Rios |
|---|
Chimeric antigen receptor (CAR) T-cell technology involves transducing targeted T cells with a T-cell receptor that enables selective targeting of tumor cells in a manner that they’re not naturally inclined to do. The use of chimeric antigen receptor (CAR) T cells for therapeutic purposes (e.g., anticancer) already is prevalent in disease research. Automation of bioprocesses using CAR T cells also has been explored. But until 2017, the connection between CAR T cells and CRISPR technology had not been established fully. One group that helped bridge those two technologies is the research team at the Memorial Sloan Kettering Institute Center for Cell Engineering. Directed by Michel Sadelain, the group has been an integral part of CAR-T therapy research, particularly targeting the CD19 molecule on white blood cells. CD19 is expressed on cells found in B-cell leukemia and resides on the cell surface, thereby making it available for antibody access. In 2017, Sadelain’s group published a study showing that directing a CD19-specific CAR to the T-cell receptor alpha constant (TRAC) locus increased T-cell potency. The modified cells “vastly outperformed conventionally generated CAR T-cells in mouse models of acute lymphoma leukemia,” with human trials planned (1). CAR T-cells typically are made using retroviruses, but they insert CAR genes into random loci in genomes. CRISPR can be used to deliver CAR genes to specified targets in genomes. And current research is exploring the use of CRISPR–Cas9 editing to create next-generation CAR T-cell products, including “universal” CAR T-cells, making these cells more potent and controllable (2). References1 Eyquem J, et al. Targeting a CAR to the TRAC Locus with CRISPR/Cas9 Enhances Tumour Rejection. Nature 543(7643) 2017: 113–117; https://doi.org/10.1038/nature21405.2 Chenggong L, Mei H, Hu Y. Applications and Explorations of CRISPR/Cas9 in CAR T-Cell Therapy. Brief. Funct. Genom. 19(3) 2020: 175–182; https://doi.org/10.1093/bfgp/elz042. |
Epigenetic Control of Therapeutic Proteins
Conferring an open chromatin state in targeted chromosome loci (DNA stretches composed of nucleosome-depleted regions) can benefit transgene expression. Indeed, cis-acting epigenetic regulatory elements can help to remodel the chromatin environment, maintaining an active transcriptional state around a transgene. One type of epigenetic regulatory element (ERE) is the scaffold/matrix attachment region (S/MAR), thanks to which recombinant proteins could improve their expression levels significantly. Another class of EREs is chromatin opening elements (UCOEs), which confer an unmethylated, open chromatin state for transgene expression. UCOEs also have helped to increase the productivity of cell line producers of recombinant proteins.
Nonetheless, S/MAR and UCOE decrease the variability of expression between the different clones. Some epigenetic elements not only can help to increase the expression level of biotherapeutics, but also can increase the number of clones that have integrated a transgene with a more defined copy number of transgenes per cell, thus accelerating the selection process.
Protein Folding and Secretion of Recombinant Cells
Secretory bottlenecks of CHO cells can be relieved by overexpressing soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs). New targets for cell-engineering approaches also can be identified based on metabolomics profiling. A bottleneck at the malate dehydrogenase II (MDHII) level was characterized for the tricarboxylic acid (TCA) cycle in CHO cells, and pyruvate metabolism was shown to vary between high-producing and low-producing anti-CD20 CHO clones (2). Over the years, many cell engineering strategies have been used in attempts to increase such titers by optimizing selection markers, gene expression, cell growth, proliferation, protein folding, and secretion. Among those engineering tools, CRISPR-Cas9 and RMCE technologies will contribute significantly to the advance of glycoprotein production shortly.
With remarkable advances in genome editing, technologies such as the CRISPR-Cas9 tool as well as the combination of next-generation sequencing with systems biotechnology exhibit extraordinary potential for discovery and modulation of novel cell engineering targets. Such efforts might finally pave the way for development of rationally designed host cells. Efforts now are ongoing to engineer PTMs in microbial, insect, and plant cell systems to make those systems more suitable for therapeutic protein production.
References
1 Baek E, Noh SM, Lee GM. Antiapoptosis Engineering for Improved Protein Production from CHO Cells. Heterologous Protein Production in CHO Cells. Humana Press: New York, NY, 2017: 71–85.
2 Lalonde ME, Durocher Y. Therapeutic Glycoprotein Production in Mammalian Cells. J. Biotechnol. 251, 2017: 128–140; https://doi.org/10.1016/j.jbiotec.2017.04.028.
3 Wang Q, et al. Glycoengineering of CHO Cells to Improve Product Quality. Heterologous Protein Production in CHO Cells. Humana Press: New York, NY, 2017, 25–44.
Dan Piestun is a biotechnology consultant and entrepreneur and was a visiting scientist at Israel’s Weizmann Institute of Science, Department of Molecular Cell Biology; [email protected]. This article has been edited with permission from the original article online at https://informaconnect.com/crispr-gene-editing-cell-line-development-engineering/?_ga=2.217983958.1382781489.1594933507-1565283457.1575662225.
You May Also Like





